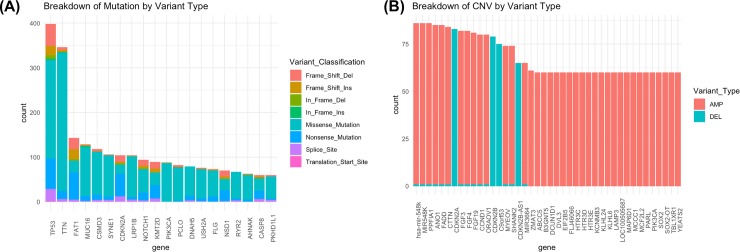
Fig 2.
A. Top 20 mutated genes in the GDC HNSCC patient cohort (N = 507). We only included variants that had high or moderate impact, which were classified as Missense, Nonsense, Nonstop, Frame Shift Deletion, Frame Shift Insertion, In Frame Deletion, In Frame Insertion, Splice Site and Translation Start Site. We excluded variants with mostly low or modifying impact, which were classified as 3’Flank, 3’UTR, 5’Flank, 5’UTR, IGR, Intron, RNA, Silent, and Splice Regions. B. Top 20 copy number altered genes in the GDC HNSCC patient cohort (N = 296). We only included copy number alterations characterized by “-2” or “+2”, for high confidence deletions and amplifications, respectively.