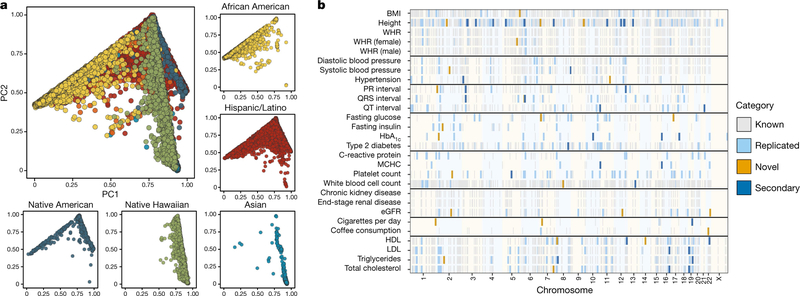
Fig. 1 |. Inclusion of multi-ethnic samples enables discovery and replication in GWAS.
a, The population substructure present in the multi-ethnic sample of PAGE (n = 49,839) revealed complex patterns preventing meaningful stratification. Here we show that PC1 and PC2 show major patterns of variation, stratified by self-identified race/ethnicity. Individuals denoted by orange self-identified as ‘Other’. b, There are 8,979 previously reported trait–variant pairs, of which 1,444 replicated at a by-trait Bonferroni-adjusted significance level for P values estimated from a Wald test in SUGEN. In addition, we found 27 novel trait–variant pairs and 38 secondary signal pairs that remained after adjusting for known variants. BMI, body-mass index; eGFR, estimated glomerular filtration rate; HbA1c, glycated haemoglobin; HDL, high-density lipoprotein; LDL, low-density lipoprotein; MCHC, mean corpuscular haemoglobin concentration; WHR, waist-to-hip ratio.