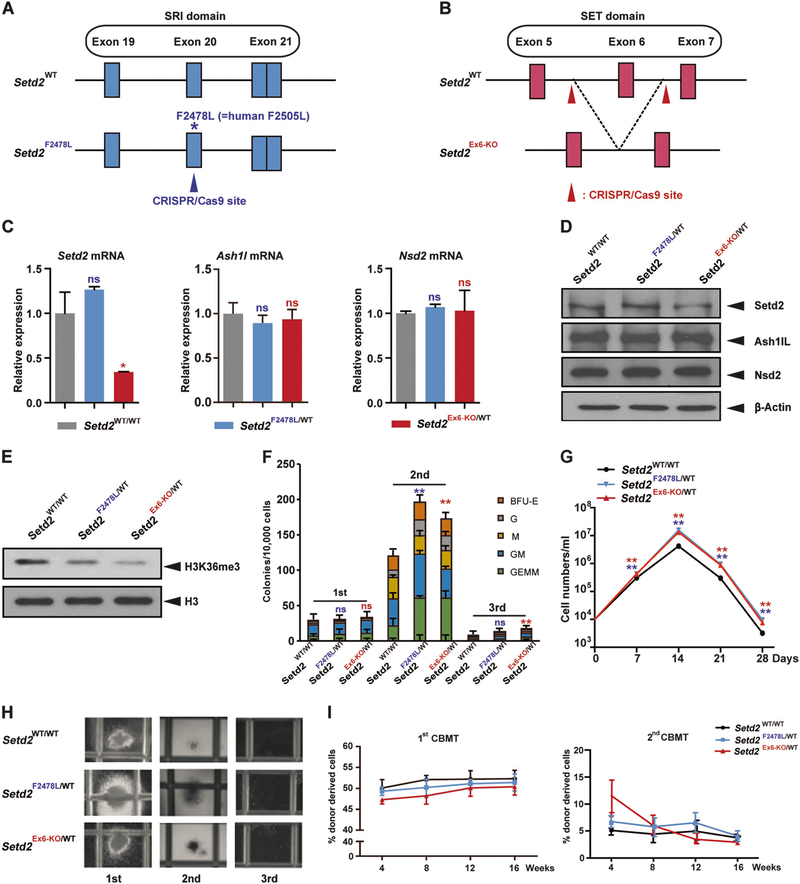
Fig. 1.
Mouse models with two distinct loss-of-function Setd2 mutations show similar phenotypes. a, b Schemes of the wild-type Setd2 locus (top) and Setd2 mutation locus (bottom) after Cas9–CRISPR- mediated modification. The Setd2F2478L point mutation locates within exon 20 in the SRI domain (a). Setd2 exon 6 locates in the SET domain (b). c Relative Setd2, Ashl1, and Nsd2 mRNA levels in c-Kit positive bone marrow cells measured by quantitative real-time PCR (Q-PCR) and normalized to β-actin levels using the Δ–CT method. d, e Endogenous Setd2, Ash1L, and Nsd2 protein expression (d), and H3K36me3 levels (e), in c-Kit positive bone marrow cells from wild type, Setd2F2478L/WT, and Setd2Ex6-KO/WT mice determined by western blot. f Bone marrow cells from wild type, Setd2F2478L/WT, and Setd2Ex6-KO/WT mice were analyzed with M3434 methylcellulose-based medium. A total of 10,000 bone marrow cells were plated in triplicate in M3434. Colony count scoring and replating was repeated every 7 days. g Cell number counts of CFUs in serial replating in (f). h Representative image of colonies in (f). i First and second competitive bone marrow transplantation. WT recipient mice were intravenously injected with 1.5 × 106 BM-MNCs from WT, Setd2F2478L/WT, or Setd2Ex6-KO/WT mice (CD45.2+) together with an equal number of CD45.1+ competitor cells. Peripheral blood cells were collected from recipients monthly and analyzed by FACS for the presence of CD45.2+ donor-derived cells. Three biological replicates of each genotype are performed in triplicate and the data are presented as the mean ± SD values. **P < 0.01