Figure 4.
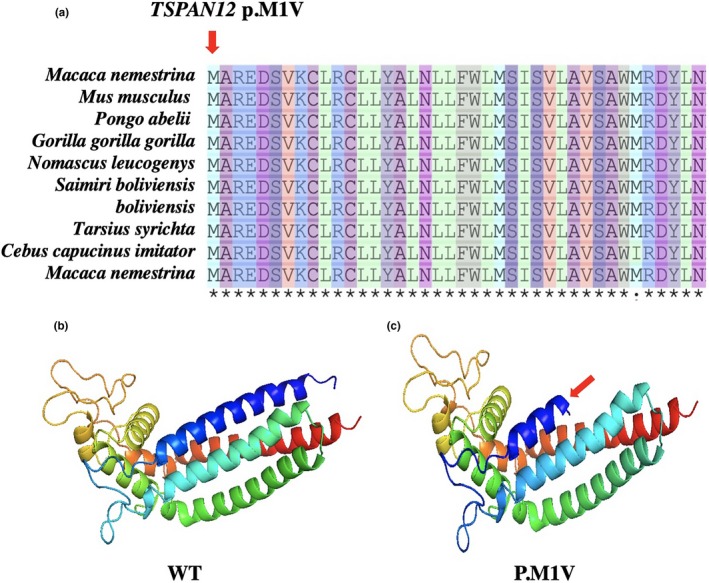
Multiple sequence alignment of start codon mutations from different species to explore the conservation of these mutations. As shown in (a), the red arrow represents mutation sites. To evaluate the influences of mutations in tetraspanin‐12, the simulation program SWISS‐MODEL was used to predict the 3‐D structure of both the mutant and wild‐type proteins. As shown in (b and c), we observed a difference between the structure of the mutated and wild‐type in tetraspanin‐12, red arrow indicates a truncated protein structure change, which resulted in a functional abnormality change
