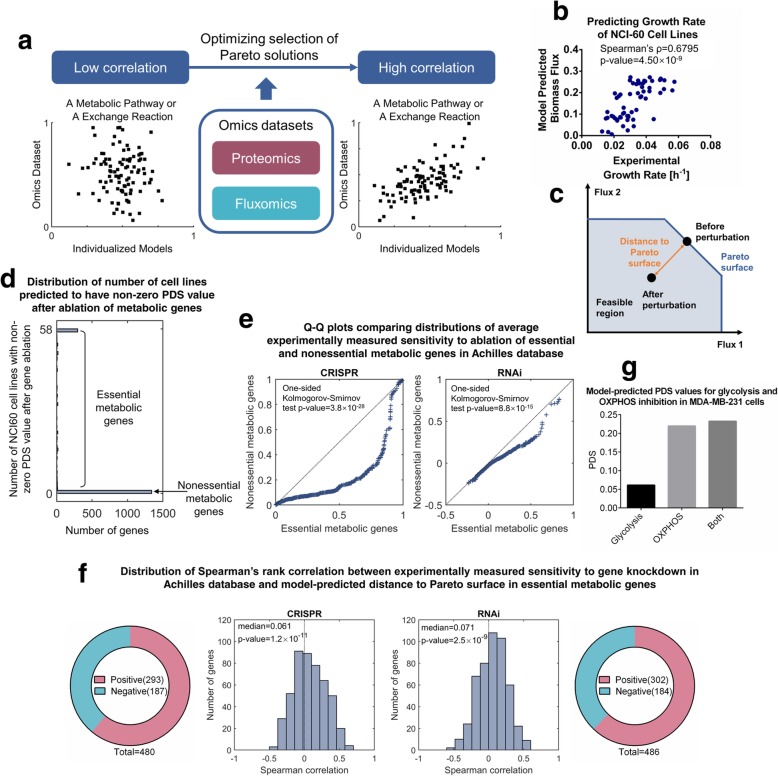
Fig. 2.
Pareto models accurately predict metabolic phenotypes of cancer cells. (a) Illustration of the strategy used in constructing the cell line-specific models based on multiple omics datasets. (b) Comparison between actual and model-predicted cell growth rates in the NCI-60 cancer cell panel. The p-value was computed using permutation test. (c) Illustration of Pareto deviation score (PDS) as a metric quantifying the impact of metabolic perturbation on cell viability. (d) Distribution of number of NCI-60 cell lines with non-zero PDS values after gene ablation in metabolic genes. (e) Quantile-quantile (Q-Q) plots comparing distributions of experimentally measured sensitivity to gene ablations between essential and nonessential metabolic genes. Left panel: CRISPR-based dataset; right panel: RNAi-based dataset. P-values were computed using one-sided Kolmogorov-Smirnov test. (f) Distributions of Spearman’s rank correlation coefficients between experimentally measured sensitivity to gene ablations and model-predicted PDS values in essential metabolic genes. P-values were computed using one-sided Wilcoxon’s signed rank test. Left panel: CRISPR-based dataset; right panel: RNAi-based dataset