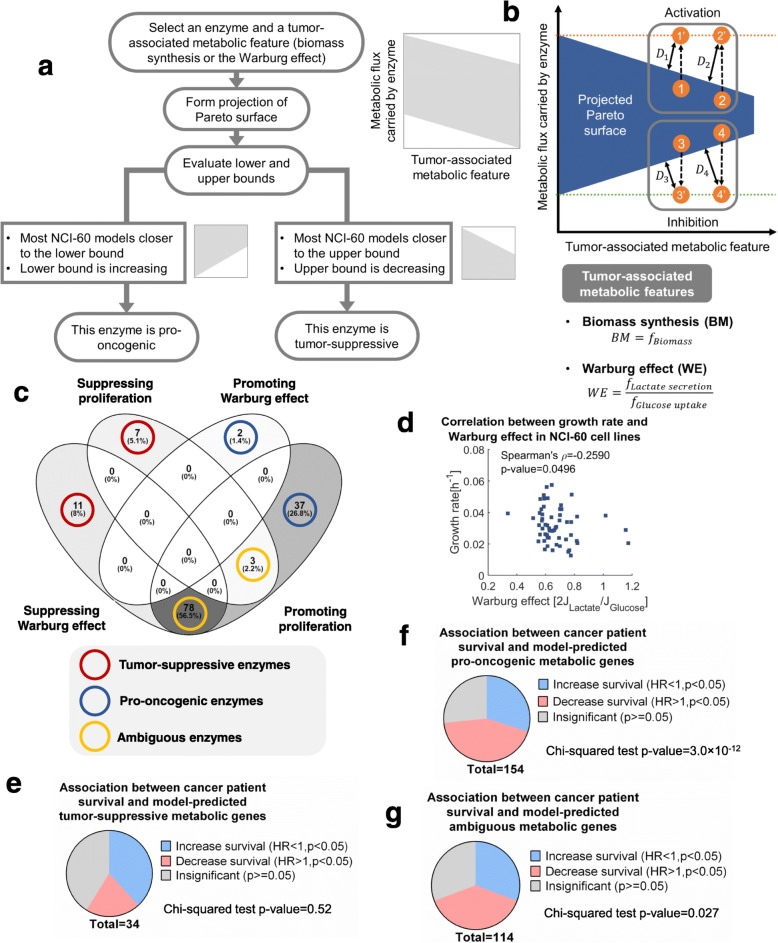
Fig. 3.
Metabolic targets identified by Pareto surface analysis correlate with cancer progression and patient prognosis. (a) Workflow of identifying potential metabolic targets essential for cell proliferation and the Warburg effect. (b) Illustration of the criteria for target identification. (c) Venn diagram showing the overlap between model-predicted proliferation-suppressing, proliferation-promoting, Warburg effect-suppressing and Warburg effect-promoting enzymes. (d) Correlation between growth rate and the Warburg effect in NCI-60 cell lines. The p-value was computed using permutation test. (e) Fraction of genes with different relationships to breast cancer patient survival in model-predicted tumor-suppressive metabolic genes. (f) Same as in (e) but for model-predicted pro-oncogenic metabolic genes. (g) Same as in (e) cmetabolic genes