Abstract
Cardiomyopathy syndrome (CMS) caused by piscine myocarditis virus is a major disease affecting the Norwegian Atlantic salmon industry. Three different populations of Atlantic salmon from the Mowi breeding program were used in this study. The first 2 populations (population 1 and 2) were naturally infected in a field outbreak, while the third population (population 3) went through a controlled challenged test. The aim of the study was to estimate the heritability, the genetic correlation between populations and perform genome-wide association analysis for resistance to this disease. Survival data from population 1 and 2 and heart atrium histology score data from population 3 was analyzed. A total of 571, 4312, and 901 fish from population 1, 2, and 3, respectively were genotyped with a noncommercial 55,735 Affymetrix marker panel. Genomic heritability ranged from 0.12 to 0.46 and the highest estimate was obtained from the challenge test dataset. The genetic correlation between populations was moderate (0.51–0.61). Two chromosomal regions (SSA27 and SSA12) contained single nucleotide polymorphisms associated with resistance to CMS. The highest association signal (P = 6.9751 × 10−27) was found on chromosome 27. Four genes with functional roles affecting viral resistance (magi1, pi4kb, bnip2, and ha1f) were found to map closely to the identified quantitative trait loci (QTLs). In conclusion, genetic variation for resistance to CMS was observed in all 3 populations. Two important quantitative trait loci were detected which together explain half of the total genetic variance, suggesting strong potential application for marker-assisted selection and genomic predictions to improve CMS resistance.
Keywords: Atlantic salmon, cardiomyopathy syndrome, field outbreak, genetic correlations, heritability, QTL analysis
One of the major diseases in Norway that affects Atlantic salmon during the seawater phase is cardiomyopathy syndrome (CMS). In 2017, the disease was reported at about 100 farms (out of ~1000 farms) along the coast of Norway (Garseth et al. 2018; Hjeltnes et al. 2018). In most cases, no clinical signs are observed until the onset of mortality in relatively large fish (~3.6 kg) several months after sea transfer. This leads to very large economic losses and reduced welfare of the fish (Løvoll et al. 2010). Moreover, it has also been observed that the presence of the disease results in large secondary losses (mortality) when the fish undergoes minor stresses like treatment for sea lice infestation (Garseth et al. 2018). Cardiomyopathy syndrome is caused by the piscine myocarditis virus (PMCV) (Haugland et al. 2011) and the disease is diagnosed by histopathology of the heart which shows moderate to severe inflammation, mostly limited to the endocardium and spongy myocardium in the atrium and ventricle (Rodger et al. 2014). In addition, real-time PCR analysis of the heart, kidney, and spleen can be used to confirm the presence of the virus (Timmerhaus et al. 2012).
Because there are no treatments or vaccines against the PMCV virus, husbandry practices (such as minimizing stressful management routines, e.g., lice and amoebic gill disease treatment) and some biosecurity measures are used to try to limit outbreaks and losses resulting from CMS (Garseth et al. 2018). An additional strategy is to increase the host resistance to the virus through selective breeding. Although the expected response per generation may be relatively small, the change in the host response is cumulative and permanent. In 2012, it was reported that there was variation between families for survival to CMS (AquaGen AS, 2015); however, the estimate of heritability was not reported. Recently, using survival data from a field outbreak of CMS, Vassgård (2017) reported heritability (liability scale) of 0.25 ± 0.04. Several authors have also reported medium to relatively high heritability for other disease resistance traits in Atlantic salmon (reviewed by Ødegård et al. 2011; Yáñez et al. 2014). When additive genetic variance for the trait is significant, it is possible to use genetic selection to increase the resistance of Atlantic salmon to diseases. For instance, selection for increased resistance to IPN in Atlantic salmon has resulted in considerable reduction in the number of outbreaks and economic losses (Hjeltnes et al. 2018).
Controlled experimental challenge tests have been commonly used to assess disease resistance phenotypes for Atlantic salmon. But challenge tests require specialized dedicated facilities and can be expensive to perform. Numerous field outbreaks occur for many diseases such as CMS in any 1 year. The main challenge with the use of field outbreak data for estimating breeding values for disease resistance is that outbreak circumstances vary and the usefulness/quality of data collected will depend on the relationship of affected animals to the breeding population, the virulence of the disease strain, environmental conditions affecting spread of the disease, and susceptibility of the fish and the ability of the farm affected to detect the occurrence of the outbreak and to collect appropriate samples. Nonetheless, field outbreak data could potentially provide useful information about the resistance of the animal to infection under grow-out conditions on-farm, which could be quite different to the situation in tanks when fish are experimentally challenged using different modes of infection and doses of disease.
Over the last decade, large genomic resources for Atlantic salmon have been developed. These resources have been used to map several quantitative trait loci (QTL), improve our understanding of the underlying genetic basis of economically important traits and provide tools for more accurate and efficient selection of fish in breeding programs. Intensive efforts have been undertaken to discover the underlying genetic architecture of disease traits in Atlantic salmon such as infectious pancreas necrosis (Houston et al. 2008; Moen et al. 2009), pancreas disease (Gonen et al. 2015), amoebic gill disease (Robledo et al. 2018), and sea lice (Tsai et al. 2016). In addition to QTL mapping efforts and marker-assisted selection approaches, genomic selection (GS) approaches have been adopted to accelerate genetic progress, providing higher selection intensity and accuracy than family pedigree-based breeding programs. Although GS in family-based Atlantic salmon breeding programs is in its early phase (2 generations of selection), initial results from several breeding programs suggests positive gains for the selected traits (Ødegard et al. 2014; Tsai et al. 2016, 2015; Bangera et al. 2017; Correa et al. 2017; Robledo et al. 2018).
Therefore, the aims of this study were to: 1) estimate and compare the heritability of resistance to CMS in populations of Atlantic salmon that were infected by a natural outbreak and by experimental challenge test, 2) detect QTLs that could potentially be utilized for marker-assisted selection to improve resistance to CMS, and 3) identify genes closely mapped to QTL for CMS resistance.
Materials and Methods
Populations
Three different populations of fish were obtained from the Mowi breeding program and used in this study. The first population was 1) a commercial grow-out fish that came from the multiplier population and were naturally infected in the field (commercial cages). The other 2 populations were full- and half-sibs of the breeding candidates (informant) from the breeding program that were 2) naturally infected with the PMVC virus during an outbreak in the field and 3) challenge tested in a controlled facility with the PMCV virus.
Field Outbreak of a Commercial Population—Population 1
In a commercial farm with approximately 190,000 fish in a single cage at Trøndelag, Norway, veterinarians confirmed an outbreak of CMS, through clinical signs and qPCR analysis of dead fish for the PMCV virus. The fish started feeding (start-feeding) in March 2016 and were transferred to sea cages in October 2016 at Lille Tørsoy, Trøndelag, Norway. We obtained 320 dead fish during the 6-week period of the CMS outbreak which started on January 2017. The dead fish were examined by veterinarians and were confirmed to be infected with the PMCV virus using quantitative polymerase chain reaction (qPCR) analysis. In April 2017, we also sampled 300 surviving fish that were critically examined and not found to be infected with the PMCV virus after mortalities had ceased. A binary phenotype was analyzed, with dead fish coded as 0 and fish that were sampled and examined to be PMCV virus free coded as 1.
Informant Population (natural infection)—Population 2
A total of 5115 eyed-eggs generated from 173 sires and 341 dams (15 eye-eggs per family) were pooled and reared together until hatching. After hatching in April 2014, the fish started feeding (start-feeding) and were communally reared until tagging. A total of 4667 (~13 fish per family) fish were pit-tagged at an average weight of 68 g. After smoltification (a process in which the fish undergoes structural and functional transformation to prepare for the transfer from fresh water to seawater) in May 2015 (at an average weight of 97 g), the fish were transferred to sea cages at Averøy, Norway. Examination of the dead fish was performed daily, and when diseases were suspected, veterinarians determined the cause of death using clinical signs and qPCR tests. The fish were full- and half siblings of the breeding candidate that were to be raised until harvest weight and recorded for slaughter traits (e.g., harvest weight, fillet color, fillet fat, etc.), however, they were infected with CMS through a natural outbreak. The PMCV virus was first detected 10 months after sea transfer when the fish with the test being tested positive for the virus after qPCR analysis. During a routine thermo-lice treatment of the fish, large numbers of mortalities were recorded, and each dead fish was collected and examined to ascertain the cause of death. Veterinarians checked for signs of edema and fibrin in the liver and spleen as well as congestion (filled with blood and fluid) and hemorrhage of the atrium. The mortalities in the entire seawater phase until slaughter were categorized as 1) unknown causes and 2) CMS-related mortalities. The total number of fish that died up until harvest (4.1 ± 1.1 kg) was 1190 (unknown = 776; CMS = 414). When analyzing this dataset, we coded the 414 fish that died with symptoms of CMS as 0; the 776 fish with unknown cause of mortality as missing and the 3477 (= 4667–1190) slaughtered fish as 1 (i.e., survivors to the PMCV virus).
Informant Population (challenge tested)—Population 3
A total of 1095 eyed-eggs were generated from 32 sires and 73 dams (15 eye-eggs per full-sibling family) and were challenged with the PMCV virus. The mating ratio consisted of 1 male to 2 females (65%) and 1 male to 3 females (35%). After hatching in April 2016, the fish were start fed and communally reared. In April 2017, the fish were smoltified before being transported from Mowi facilities at Øyerhamn, Norway to VESO Vikan, Namsos, Norway for the challenge test in May 2017. After 2 weeks of acclimatization, they were challenged tested through an intraperitoneal injection of the PMCV virus that originated from a highly virulent field outbreak of CMS, which was confirmed by qPCR analysis. At the end of the challenge period (9 weeks), heart tissues were obtained from each fish for histopathology analysis was undertaken as described under population 2. Histopathology of heart tissues was undertaken by the FishVet Group, Norway (http://fishvetgroup.no/en/). Both the atrium and ventricular tissue was examined for damage using the scoring system described in Table 1, with a score of 0 representing no visible lesions to a score of 3 which represented >50% of the heart tissue showing confluent lesions. Only histology scores of the atrium were used as the phenotype in this population because we obtained similar results using ventricular histology scores.
Table 1.
Scoring scheme for the histology of heart (atrium) tissue affected by cardiomyopathy syndrome
| Score | Description |
|---|---|
| 0 | No histopathological findings |
| 1 | Few (<7) focal lesions |
| 2 | Several distinct lesions and increased mononuclear infiltration |
| 3 | Multifocal to confluent lesions in >50% of tissues + moderate – severe leukocyte infiltration |
Genotyping and Quality Control
DNA was extracted from fin clips of all the fish using commercial kits by IdentiGEN, Ireland.). A total of 5940 samples (620, 4350, and 985 from population 1, 2, and 3, respectively) were genotyped with a ThermoFisher Axiom 57K single nucleotide polymorphism (SNP) array (NOFSAL03, 55735 markers) that was developed by Nofima in collaboration with Mowi and SalmoBreed. Genotyped samples were quality checked within population with PLINKv1.9 (Chang et al. 2015) using the following procedure: Samples and SNPs with a call rate <95% were discarded. SNPs with Hardy Weinberg P-value (Fishers exact test) <10–15 and those with minor allele frequency <2% were removed. Samples that were <25% and >47% heterozygous were discarded. After genotype quality checks the total number of samples (markers in bracket) that were used for further analysis were 571 (54498), 4312 (53738), and 901 (53678) for population 1, 2, and 3, respectively.
Statistical Analysis
(Co)variance Estimations
Variance and covariance components were estimated with genomic relationships instead of pedigree information to be able to estimate the genetic correlations between the 3 populations. The following linear mixed animal model was applied:
where (binary – dead = 0 and survived = 1), (binary – dead = 0 and survived = 1) and (histology scores ranged from 0 to 3) are vectors of phenotypes for the animals in population 1, 2, and 3, respectively; , , and are incidence matrices relating the phenotypic records to their respective mean (, , and ) of the 3 population; , , and are incidence matrices linking the phenotypes in each population to their genomic breeding values (, and ); , and are vectors containing the random residual effects for the 3 populations. Genomic breeding values were assumed to follow a multivariate normal distribution , where , and are the genetic variances for population 1, 2, and 3, and , , and were the genetic covariances between the populations. The multipopulation genomic relationship matrix () was computed following the approach described by Wientjes et al. (2017) as:
where , , and contains the centered genotypes of population 1, 2, and 3. The marker genotypes coded as 0 (AA), 1 (AB | BA) and 2 (BB) were centered with the observed allele frequency of each population, where (), (), and () are the allele frequencies of marker for population 1, 2, and 3, respectively. Residual effects were assumed to be normally distributed but with no residual covariance between the populations such that , where was an identity matrix and , , and were the residual variances for population 1, 2, and 3. The genetic and residual (co)variances were estimated in ASREML v4 software (Gilmour et al. 2009).
Marker Association Analysis
Genome-wide association analysis was performed using a linear mixed animal model approach for each of the population separately. To account for genetic stratification (Supplementary Figure 1) within each of the populations, the first 5 eigenvectors from a principal component analysis (Chang et al. 2015) were used as covariates. The following model was used
where are either the binary phenotypes for population 1 and 2 or the histology scores for population 3, is the overall mean are the first 5 eigenvectors, is the incidence matrix for marker containing marker genotypes, is the allele substitution effect of marker , was the incidence matrix linking phenotypes to their genomic breeding values () and was assumed to follow normal distribution , where, is the genomic relationship matrix constructed for each population and was equivalent to the diagonal block of the matrix, and is the vector of random residual effects which followed a normal distribution , where, was an identity matrix and was the residual variance. The analysis was undertaken with GCTA (Yang et al. 2011a) using the “--mlma-loco” approach which ensures that the effect of marker is estimated by accounting for the additive genetic variance () and relationships captured by chromosomes other than the one the marker is located. Markers were considered significant when they exceeded the Bonferroni threshold for multiple testing of 0.05 divided by the number of markers that passed the quality check for that population. To quantify the level of inflation (lambda) of the observed -values compared to the expected -values, we computed lambda () as (Utsunomiya et al. 2013) and the results were presented in a quantile–quantile (Q–Q) plots. The proportion of total genetic and phenotypic variance explained by significant markers was computed from the estimated allele substitution effects () and allele frequencies of each marker as: and.
Bioinformatics and Candidate Genes
SNPs passing a genome-wide significance threshold () were further examined to identify genes potentially involved in CMS resistance. Bedtools (v2.26.0) was used to identify the SNP position relative to the closest gene or genes. This was conducted by making a bed file from the SNP positions and using intersectBed with the Atlantic salmon genome version 2.2 (https://www.ncbi.nlm.nih.gov/assembly/GCF_000233375.1/). Further investigation of the genes involved was conducted using NCBI resources (BLAST, Gene, and Pubmed) and the potential effect of SNPs within exons was assessed by translating each SNP variant using ExPASy Translate tool (https://web.expasy.org/translate/) and the predicted protein sequences from NCBI.
Results
Phenotypic Distribution
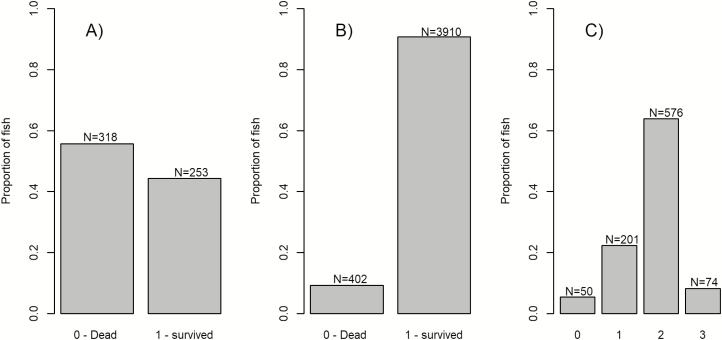
To maximize power to detect possible QTLs associated with resistance to the PMCV virus, we sampled approximately equal numbers of dead and surviving fish from population 1, however, after genotype quality checks, the percentage of dead fish was slightly higher (55.7%) than the surviving fish that were sampled (44.3%, Figure 1A). For population 2 (an informant population which encountered a natural outbreak of the PMCV virus), only 402 fish (9.3%) out of 4312 died from the CMS outbreak, the rest of the population (90.7%, Figure 1B) were considered as survivors and used in the analysis. Since the PMCV challenge test model did not result in mortalities, we performed histology of the heart (atrium) for all the challenge tested fish. The mean histology score was 1.750 (SD = 0.680), however, 86.2% of all fish had either a score of 1 (22.3% with no histopathological findings) or a score of 2 (63.9% with few (<7) focal lesions, Figure 1C).
Figure 1.
Distribution of traits in the 3 population. (A) Binary phenotype (dead or survived) recorded in population 1, (B) binary phenotype (dead or survived) recorded in population 2, and (C) heart tissue (atrium) histology scores (0 to 3) recorded in population 3.
Heritability and Genetic Correlation Between the Populations
Heritability estimates for survival to PMCV in population 1 and 2 were 0.383 ± 0.076 and 0.117 ± 0.018, respectively (Table 2). A higher heritability (0.464 ± 0.055) was obtained for histopathological score of the atrium in the controlled challenge test of population 3 (Table 2).
Table 2.
Variance components and heritability estimate (standard error; in bracket) for the 3 populations
| Parameters | Field outbreak | Challenge test Population 3 | |
|---|---|---|---|
| Productiown population 1 | Informant population 2 | ||
| 0.097 (0.023) | 0.010 (0.002) | 0.226 (0.038) | |
| 0.157 (0.017) | 0.076 (0.002) | 0.262 (0.020) | |
| 0.254 (0.017) | 0.086 (0.002) | 0.488 (0.031) | |
| 0.383 (0.076) | 0.117 (0.018) | 0.464 (0.055) |
, genetic variance; , residual variance; , phenotypic variance; , heritability; binary phenotype (dead = 0 and survived = 1) was used in population 1 and 2, histology score of the heart (atrium) was the recorded phenotype in population 3.
The estimated genetic correlation (using the genomic relationship matrix) between survival to PMCV in population 1 and population 2 was moderate (0.609 ± 0.225; Table 3). The genetic correlation between survival (population 1 and 2) and histology scores (population 3) was favorable and ranged from −0.514 ± 0.190 to −0.593 ± 0.265 (Table 3). The estimates of heritability and genetic correlations between the populations were all significantly different from zero (P<0.05) based on a likelihood ratio test (results not presented).
Table 3.
Genetic correlations (standard error) between the binary survival traits (Production and Informant populations) and the atrium-score trait (Challenge test population)
| Populations | 1 Production |
2 Informant |
|
|---|---|---|---|
| 1 | Production | ||
| 2 | Informant | 0.611 (0.225) | |
| 3 | Challenge test | −0.593 (0.262) | −0.514 (0.190) |
Marker Association
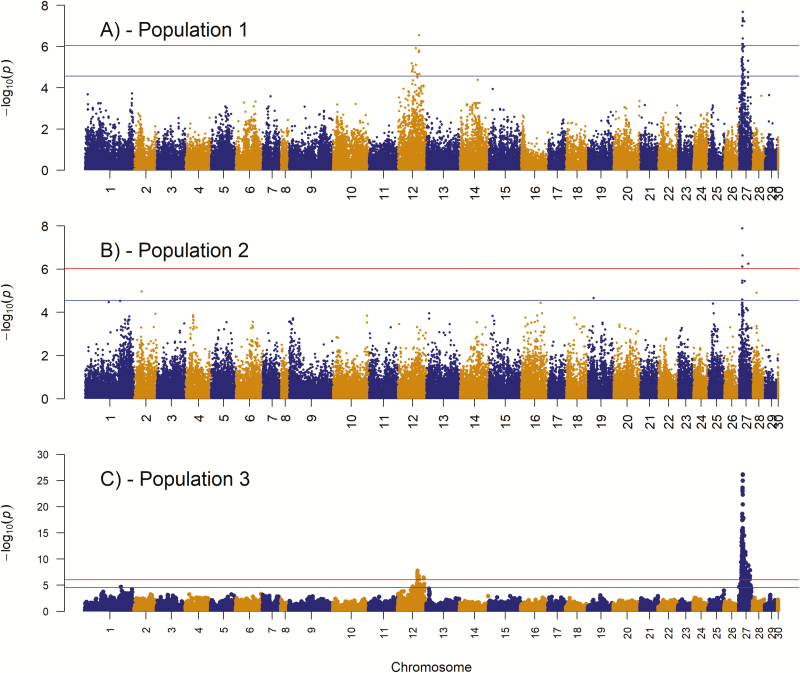
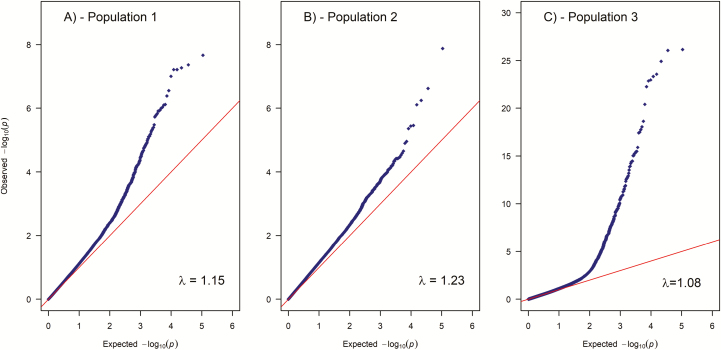
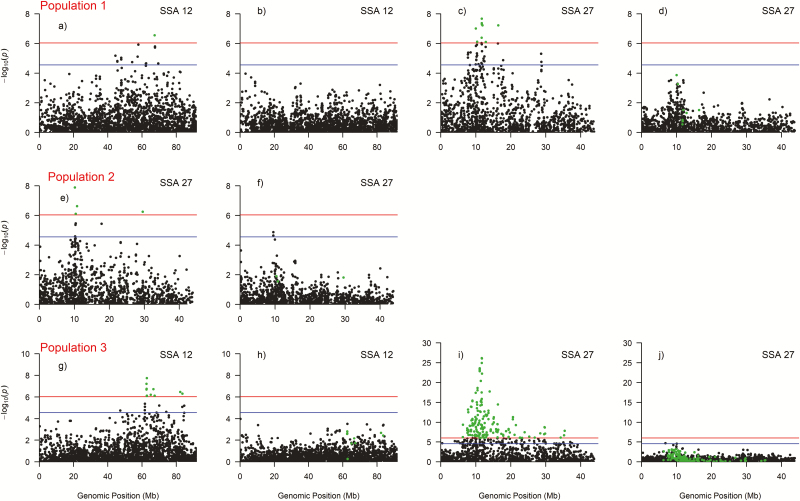
We detected 24 and 186 markers on chromosome 12 and 27, respectively that exceeded the genome-wide Bonferroni significance level (; Figure 2 and Supplementary File 2). The putative QTL on chromosome 12 was present in population 1 and 3 but not in population 2, while the QTL on chromosome 27 was present in all 3 populations (Figure 2A–C). The observed P-values were inflated ( of ~1.1 indicates relatively good concordance between observed and the assumed distributions of the test statistic) with lambda values ranging from 1.08 to 1.23 (Figure 3). When the most significant SNP was included as a covariate, one at a time for each QTL region and population, to investigate the possibility of multiple QTLs in any of the regions on chromosome 12 and 27, none of the surrounding SNPs showed associations with the trait (Figure 4b,d,f,h, and j).
Figure 2.
Manhattan plot of resistance to cardiomyopathy syndrome (CMS). (A) Binary phenotype recorded from field outbreak on the commercial farm population 1. (B) Binary phenotype recorded from field outbreak of the informant pedigreed population 2. (C) Histology score of heart tissue (atrium) based on challenge test information of the informant pedigreed population 3. Red and blue horizontal line represent genome-wide significant threshold () and chromosomal-wide significant threshold for the population (), respectively. See online version for full colors.
Figure 3.
Quantile–quantile plot of values for each of the 3 populations. See online version for full colors.
Figure 4.
Manhattan plots of chromosome 12 and 27 for the 3 populations; (a), (b), (c), and (d) are plots from population 1; (e) and (f) are plots from population 2; and (g), (h), (i), and (j) are plots from population 3. Plots (b), (d), (f), (h), and (j) were obtained by fitting the top most significant marker as covariate in the genome-wide association analysis. Red and blue horizontal line represent the genome-wide significant threshold () and the chromosomal-wide significant threshold for the population (), respectively. See online version for full colors.
For population 1, 1 marker on chromosome 12 (located at 67,351,339 bps) and 9 markers on chromosome 27 (spanned from 10,023,660 to 16,436,601 bps) exceeded the genome-wide threshold (Figures 2 and 4). The most significant marker for population 1 on chromosome 27 was located at 11,724,500 bps (Table 4). The frequency of the favorable allele for the top significant marker for chromosome 12 and 27 were 0.531 and 0.253, with each marker explaining about 12% and 14% of the total genetic variance, respectively.
Table 4.
Summary statistics of the most significant markers for the 3 populations
| Populations | CHR | Position (bp) | SNP | A1 | A2 | Freq (A1) | Beta (A1) | P-value | Vg1 | Vp2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Production | 12 | 67351339 | AX-96382208 | B | A | 0.469 | −0.159 | 2.81E−07 | 0.120 | 0.049 |
| 27 | 11724500 | AX-96420652 | B | A | 0.253 | 0.197 | 2.15E−08 | 0.140 | 0.057 | ||
| 2 | Informant | 27 | 10160666 | AX-86985828 | A | B | 0.157 | −0.055 | 1.32E−08 | 0.079 | 0.009 |
| 3 | Challenge test | 12 | 62828648 | AX-96152485 | B | A | 0.412 | 0.216 | 1.79E−08 | 0.101 | 0.046 |
| 27 | 11723738 | AX-97886034 | A | B | 0.397 | −0.384 | 6.98E−27 | 0.317 | 0.145 |
1Proportion of the genetic (Vg) and 2phenotypic (Vp) variance explained by the most significant markers.
For population 2, 4 markers on chromosome 27 were significant, while none of the markers on chromosome 12 was significant (Figures 2 and 4). Three of these markers spanned from 10,160,666 to 10,781,998 bps (a region of 621,332 bps), while the other marker was located at 29,548,089 bps. The most significant marker (P = 1.322 × 10−8) was located at 10,160,666 bps and it explained 7.9% of the total genetic variance.
For population 3, the QTL peaks on chromosome 12 and 27 were very strong (Figures 2 and 4), with 175 markers exceeding the Bonferroni threshold. Out of the 175 significant markers, 11 and 164 markers were on chromosome 12 and 27, respectively. The most significant marker (P = 6.9751 × 10−27) for this population was also on chromosome 27 and was located at 11,723,738 bps. The frequency of the favorable allele was 0.397 and it explained 31.7% and 14.5% of the total genetic and phenotypic variance. The entire region on chromosome 27 spanned from 6,292,857 to 35,401,496 bps. Six out of the 11 markers on chromosome 12 spanned a region of 236,290 bps (between 62,592,358 and 62,828,648 bps) and the most significant marker (P = 1.7943 × 10−8 and at 62,828,648 bps) was located within this region. This marker explained 10.1% of the total genetic variance (Table 4).
Comparing the most significant marker from one population to the other, we observed a consistent direction (positive or negative) of the allele substitution effect in all the populations, although in some cases the marker was not significant () (Table 5).
Table 5.
Summary statistics of the 5 significant markers in all the 3 population
| Chromosome | SNP | Position(bps) | A1 | A2 | Freq (A1) | beta (A1) | P-value | Population1 | Population marker was significant |
|---|---|---|---|---|---|---|---|---|---|
| 27 | AX-86985828 | 10160666 | A | B | 0.157 | −0.055 | 1.32E−08 | Informant | informant |
| 27 | AX-86985828 | 10160666 | A | B | 0.383 | −0.145 | 1.26E−05 | Production | informant |
| 27 | AX-86985828 | 10160666 | A | B | 0.141 | 0.110 | 2.28E−02 | Challenge test | informant |
| 12 | AX-96382208 | 67351339 | B | A | 0.542 | −0.003 | 6.82E−01 | Informant | Production |
| 12 | AX-96382208 | 67351339 | B | A | 0.469 | −0.159 | 2.81E−07 | Production | Production |
| 12 | AX-96382208 | 67351339 | B | A | 0.263 | 0.213 | 7.69E-07 | Challenge test | Production |
| 27 | AX-96420652 | 11724500 | A | B | 0.382 | −0.009 | 2.28E−01 | Informant | Production |
| 27 | AX-96420652 | 11724500 | A | B | 0.747 | −0.197 | 2.15E−08 | Production | Production |
| 27 | AX-96420652 | 11724500 | A | B | 0.329 | 0.293 | 1.26E−16 | Challenge test | Production |
| 12 | AX-96152485 | 62828648 | A | B | 0.289 | 0.006 | 3.98E−01 | Informant | Challenge test |
| 12 | AX-96152485 | 62828648 | A | B | 0.359 | 0.045 | 1.59E−01 | Production | Challenge test |
| 12 | AX-96152485 | 62828648 | A | B | 0.587 | −0.216 | 1.79E−08 | Challenge test | Challenge test |
| 27 | AX-97886034 | 11723738 | A | B | 0.310 | 0.021 | 1.06E−02 | Informant | Challenge test |
| 27 | AX-97886034 | 11723738 | A | B | 0.132 | 0.235 | 4.29E−08 | Production | Challenge test |
| 27 | AX-97886034 | 11723738 | A | B | 0.397 | −0.384 | 6.98E−27 | Challenge test | Challenge test |
1Binary phenotype (dead = 0 and survived = 1) was used in population 1 (Production) and 2 (informant), histology score of the heart (atrium) was the recorded phenotype in population 3 (Challenge test).
Most of the significant SNPs were located within genes, including UTRs, introns, exons, and intergenic regions (Supplementary File 2). Thirty-six of the SNP markers resided within exons and 18 of them would cause amino acid changes. Genes located within ± 50 kbs of the top significant markers obtained from the association analysis of the 3 populations were investigated. In all, 4 distinct 50 kb regions around the most significant markers were investigated (Table 6). On chromosome 12, 2 QTL regions (region 12_1: 62,778,648 to 62,878,648 bps and region 12_2: 67,301,339 to 67,401,339 bps) were defined based on the top significant SNP that was obtained in population 1 and 3. Two QTL regions were also defined for chromosome 27; region 27_1 and 27_2 spanned from 10,110,666 to 10,210,666 bps (population 2) and 11,674,500 to 11,774,500 bps (population 1 and 3), respectively.
Table 6.
Genes located close to the top significant markers of chromosome 12 and 27 for the 3 population
| Chromosome | QTL region1 | Population | Gene | Gene name | ||
|---|---|---|---|---|---|---|
| Left position (bp) | Right position (bp) | |||||
| 12 | SSA12_1 | 62,778,648 | 62,878,648 | Challenge test | LOC106565906 | Draxin-A |
| LOC106565953 | Uncharacterized LOC106565953 | |||||
| LOC106565907 | Pantothenate kinase 4-like (PANK4) | |||||
| LOC100353126 | Angiotensin II receptor associated protein (AGTRAP) | |||||
| SSA12_2 | 67,301,339 | 67,401,339 | Informant | LOC106565841 | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1-like ((MAGI1) | |
| 27 | SSA27_1 | 10,110,666 | 10,210,666 | Informant | LOC100136924 | Transport-associated protein 2b (TAP2B) |
| LOC106588399 | Proteasome subunit beta type-6-a like protein-like (PSMB6-A) | |||||
| LOC100136936 | Proteasome subunit beta type-9a (PSMB9-A) | |||||
| LOC106588263 | Uncharacterized LOC106588263 | |||||
| LOC106588398 | Proteasome subunit beta type-7-like | |||||
| LOC106455085 | Transfer RNA valine (anticodon GAC) (TRNAV-GAC) | |||||
| LOC106588400 | Proteasome subunit beta type-8-like | |||||
| LOC106588401 | Class I histocompatibility antigen, F10 alpha chain-like (HFA1) | |||||
| SSA27_2 | 11,674,500 | 11,774,500 | Challenge test and Production | LOC106588340 | Phosphatidylinositol 4-kinase beta (PI4KB) | |
| LOC106588342 | Protein prune homolog | |||||
| LOC106588341 | Protein FAM63A-like | |||||
| LOC106588344 | Guanine nucleotide-binding protein G(s) subunit alpha pseudogene (GNAS) | |||||
| LOC106588343 | BCL2/adenovirus E1B 19 kDa protein-interacting protein 2-like (BNIP2) | |||||
| LOC106588345 | CDC42 small effector protein 1-like |
1QTL region was defined as ± 50 kbs to the left and right of the most significant SNP in each population.
Several interesting genes with functions known to affect viral resistance were found to map to the QTL regions (Table 6). The magi1 gene (membrane-associated guanylate kinase, WW, and PDZ domain containing 1) was the only gene in region 12_2. The class I histocompatibility antigen, F10 alpha chain-like (ha1f) gene was found in region 27_1. Genes in region 27_2 included the transcript for phosphatidylinositol 4-kinase beta (pi4kb) and BCL2/adenovirus E1B 19 kDa protein-interacting protein 2-like (bnip2).
Discussion
The main aim of this study was to estimate heritability and to detect QTLs for CMS resistance in 3 different year-class populations of Atlantic salmon. The recorded data used was binary survival data from field outbreaks (2 of the populations) and atrium-score data from a challenge test (the third population). In addition, we estimated the magnitude of the genetic correlations between the 3 mentioned traits in these populations. We obtained low to high heritability estimates (0.12–0.46) with survival data from field outbreaks and challenge test experiments. A moderate and favorable genetic correlation was obtained between the traits from the 3 populations. The genome-wide association study identified a putative QTL on chromosome 27 in all 3 populations and on chromosome 12 in 2 of the populations. Both QTLs explained about 7.6% to 31.7% of the total genetic variance and about 1% to 14.5% of the phenotypic variance. Functionally interesting candidate causative genes (hfa1 and bnip2) were found to map near to the SNPs that were most significant.
Heritability of Resistance to CMS Infection
Low to moderate heritability was estimated for survival (0.38 ± 0.08 for population 1 and 0.12 ± 0.02 for population 2) while higher heritability was estimated for the atrium-score trait (0.46 ± 0.06). The higher heritability estimates for survival in population 1 may be due to the much higher mortality (55.7%) than in population 2 (9.3%). When the 2 binary heritability estimates were transformed to the underlying liability scale according to Dempster and Lerner (1950), heritability estimate were 0.61 for population 1 and 0.36 for population 2. The lower heritability estimates in population 2 may be due to a relatively higher number of noninformative (zero mortality) families (Ødegård et al. 2007) as well as the different sampling procedures used to collect data from the 2 populations. While those of population 2 were the survivors of the entire seawater period, the 300 survivors of population 1 were sampled and critically examined not to be infected with the PMCV virus after an equal number of dead fish had been recorded during a 6 weeks CMS outbreak. The heritability estimates for survival in population 1 are likely to be more specific estimates of the susceptibility to CMS than those for survival in population 2. In addition, some of the mortalities recorded in population 2 could have been due to causes other than CMS. This might be part of the reason for the relatively moderate genetic correlation (0.61 ± 0.23) detected between survival to CMS in the 2 populations. For this reason, the sampling strategy and the survival data from population 1 probably yields more power for mapping QTL affecting the susceptibility to CMS than the survival data from population 2. Nonetheless, the magnitude of the heritability estimates in all 3 populations confirm that there is substantial genetic variation in susceptibility to CMS and that genomic or marker-assisted selection could be effectively used to improve the resistance of Atlantic salmon to the PMCV virus. Moreover, the heritability estimates in this study are of a similar level to those detected for several other disease traits in Atlantic salmon (see the review by Yáñez et al. 2014).
There are challenges with using field outbreak information from commercial populations for heritability estimation and mapping QTLs. First, it can be difficult to identify cause-specific mortalities (exact cause of death). Furthermore, without genomic information, it would not be possible to estimate heritability from commercial populations with unknown pedigree. The fish in commercial cages might have been produced from a few sires and dams, and therefore even with known pedigree, the estimate of heritability might not be very reliable. However, with genomic information, it is possible to estimate heritability from nonpedigreed populations as well as from populations with very few families and large number of offspring per family (Ødegård and Meuwissen, 2012). Although estimate of heritability from genome-wide identity-by-state markers can be biased upwards (Fernando et al. 2017). Finally, field outbreaks might not provide regular mortality information and could therefore be unreliable for routine selection of breeding candidates (Bishop et al. 2012; Yáñez et al. 2014).
Genetic Correlations Between Traits from the Different Populations
At first glance, the favorable but moderate genetic correlation of atrium histology scores with survival due to CMS outbreak in the field (population 1 and 2) indicates that the histology score of the atrium is not a good predictor of survival to CMS in the field. However, to better estimate the genetic correlation between survival and atrium histology score, these 2 traits should be measured on the same individuals or siblings, so that the magnitude of the correlation is not influenced by relatedness between independent populations which was the case in this study.
The moderate genetic correlation observed between populations may also be because the populations are different year-classes of Atlantic salmon established from different genetic bases during the late 1960’s and early 1970’s with little flow of genetic material and relatively low genetic connectedness between the 2 populations. This explains the population stratification that was evident from the principal component analysis (Supplementary File 1). Furthermore, a moderate genetic correlation between populations may be obtained because 1) linkage disequilibrium between markers and QTL could be different between populations and 2) markers associated with the QTL could differ in allele frequency between the populations. The magnitude of the genetic correlations between populations for a specific trait also reflects the expected genomic prediction potential across populations. (Raymond et al. 2018) reported that accuracies were 50% less when the genetic correlation between 2 breeds was 0.5 instead of 1. In this study, we computed the genetic correlations using all markers, however, since the LD phase between QTL and markers could be more persistent at causal loci (i.e., estimated allele substitution effect are similar and in the same direction) than at noncasual loci (Wientjes et al. 2017; Raymond et al. 2018), we also investigated the effect of using only markers on chromosome 27 to construct the genomic relationship matrix and compute the genetic correlations between populations. This approach will estimate the genetic correlation between the marker effect of the 2 QTL region/chromosome between the population. The estimated genetic correlation using these 2 chromosomal regions was ~1.0 between population 1 and 2, −0.79 between population 1 and 3 and −0.59 between population 2 and 3. An increase in the estimate of genetic correlations between populations when casual variants are used compared to all loci have previously been observed in other studies (Raymond et al. 2018).
Putative QTL Positions
P-values in this study were inflated ( for population 1 and 2), however, this is to be expected for structured populations with large full- and half-sib families (Yang et al. 2011b).
We observed 2 regions on chromosome 27 that were highly associated with resistance to CMS, however, since the power to detect QTLs for resistance to CMS was much larger in population 1 and 3 than in population 2, it is more likely that the causal mutation is located close to the 11.72 Mbps region than the 10.16 Mbp region. For chromosome 12, 2 regions were identified in population 1 (62828648 bps) and 3 (67351339 bps), and these 2 regions were 4.5 Mbps apart. The reasons for the discrepancies in the location of the top significant markers on chromosomes 12 and 27 are difficult to explain. For example, the top significant marker of population 1 (AX-86985828, with frequency of allele B equal to 0.16), was equally frequent in population 3 (frequency of allele B equal to 0.14), but this marker was not significant in population 3. We speculate that, the reason for these discrepancies is because the markers on this SNP array are not in complete linkage disequilibrium with the QTL. Therefore, fine mapping of these regions with sequence information should be undertaken to narrow down the QTL region and possibly identify the causative gene and the quantitative trait nucleotides.
There was consistency in the direction of the allele substitution effects in the 3 populations and this suggests that, the LD phase between the markers and the causative mutation is in the same direction in the 3 populations, however, since allele frequencies differ, the magnitude of the allele substitution effect and the proportion of variance explained by the QTL region is expected to be different in the 3 populations.
The QTL region on chromosome 27 explained ~30% of the total genetic variance (results from the challenge test). Several medium to large QTLs have also been found for several disease traits in Atlantic salmon such as pancreas disease (Gonen et al. 2015) and infectious pancreatic necrosis (Houston et al. 2008; Moen et al. 2009). Using the top marker as a covariate before computing the allele substitution effect of all other markers on chromosome 27 removed the QTL peak (Figure 4). This is an indication that, the top significant marker can be used to tag the QTL and can be used for marker-assisted selection (MAS). Therefore, classical family-based selective breeding, supplemented with MAS to capitalize on the genetic variation within families, can be used as one of the strategies to combat this disease. Application of marker-assisted selection to selective breeding utilizing marker genotypes for these large QTLs have led to substantial increases in genetic gain in some Atlantic salmon breeding programs (Storset et al. 2007).
Genes Located in the QTL Region
Several genes with functions potentially affecting viral resistance were found to map to the regions identified. In mammals, the PDZ1 and PDZ3 domains of the magi1 gene have been shown to regulate the amount of coxsackievirus and adenovirus receptor (car), which directly impacts pathogenicity of these viruses (Kolawole et al. 2012). The ha1f gene is part of the major histocompatibility complex class 1 (mr1) family of genes. The mr1 (LOC106588402) gene was only 109 kbs away from the top significant marker of population 1. Specific alleles of mhc I and II have been found to be associated with resistance to infectious salmon anemia in Atlantic salmon (Kjøglum et al. 2006; Grimholt et al. 2003). The product of pi4kb has been found to be important in viral replication for several types of viruses (Delang et al. 2012). One particular virus, Coxsackievirus, produces a similar phenotype, myocarditis, in humans, and pi4kb is associated with resistance/susceptibility to Coxsackievirus infection in humans (Dorobantu et al. 2014). The bcl2/adenovirus E1B 19 kDa protein-interacting protein 2-like (bnip2) interacts with a protein responsible for the protection of virally induced cell death, and with cdc42, a gene which was also found to map in close proximity to QTL 27_2 (Boyd et al. 1994). In the absence of E1B-19k, cellular and viral DNA degradation occurs, and the death of viral host cells is promoted, and both these actions inhibit viral replication (White, 2001). All these genes warrant further attention as potential candidates affecting viral immune function.
Conclusion
Our results from field outbreaks and challenge tests show that resistance to CMS in Atlantic salmon is heritable (0.12–0.46), and that breeding populations could be selected for improved CMS resistance to help reduce outbreaks of the disease. We also identified 2 QTL regions on chromosome 27 and 12 across all 3 populations which explained 7.6% to 31.7% of the total genetic variance. Interesting genes mapping to these QTL regions that could play a role in resistance to CMS included magi1, pi4kb, bnip2, and ha1f. These results also highlight the potential of using field outbreak information from nonpedigreed populations to estimate heritability and map QTLs in Atlantic salmon populations.
Funding
This work was partly funded by the Norwegian Research Council, project number 254880.
Data Availability
The datasets used in this study were from a private breeding company, therefore we are not permitted to share phenotypic and genotypic data directly, however, data is available upon request from the corresponding author.
Supplementary Material
Acknowledgments
We thank Mowi Genetics As for providing us with genotype data of population 2 and 3. The ASreml analysis was performed on the Abel Cluster (http://www.hpc.uio.no/).
References
- AquaGen AS 2015. Protection against CMS – Better heart health. Available from: https://aquagen.no/en/products/salmon-eggs/product-documentation/resistance-against-cms/ [Google Scholar]
- Bangera R, Correa K, Lhorente JP, Figueroa R, Yáñez JM. 2017. Genomic predictions can accelerate selection for resistance against Piscirickettsia salmonis in Atlantic salmon (Salmo salar). BMC Genomics. 18:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop SC, Doeschl-Wilson AB, Woolliams JA. 2012. Uses and implications of field disease data for livestock genomic and genetics studies. Front Genet. 3:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd JM, Malstrom S, Subramanian T, Venkatesh LK, Schaeper U, Elangovan B, D’Sa-Eipper C, Chinnadurai G. 1994. Adenovirus E1B 19 kDa and Bcl-2 proteins interact with a common set of cellular proteins. Cell. 79:341–351. [DOI] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa K, Bangera R, Figueroa R, Lhorente JP, Yáñez JM. 2017. The use of genomic information increases the accuracy of breeding value predictions for sea louse (Caligus rogercresseyi) resistance in Atlantic salmon (Salmo salar). Genet Sel Evol. 49:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delang L, Paeshuyse J, Neyts J. 2012. The role of phosphatidylinositol 4-kinases and phosphatidylinositol 4-phosphate during viral replication. Biochem Pharmacol. 84:1400–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempster ER, Lerner IM. 1950. Heritability of threshold characters. Genetics. 35:212–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorobantu CM, van der Schaar HM, Ford LA, Strating JR, Ulferts R, Fang Y, Belov G, van Kuppeveld FJ. 2014. Recruitment of PI4KIIIβ to coxsackievirus B3 replication organelles is independent of ACBD3, GBF1, and Arf1. J Virol. 88:2725–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando RL, Cheng H, Sun X, Garrick DJ. 2017. A comparison of identity-by-descent and identity-by-state matrices that are used for genetic evaluation and estimation of variance components. J Anim Breed Genet. 134:213–223. [DOI] [PubMed] [Google Scholar]
- Garseth ÅH, Fritsvold C, Svendsen JC, Bang Jensen B, Mikalsen AB. 2018. Cardiomyopathy syndrome in Atlantic salmon Salmo salar L.: a review of the current state of knowledge. J Fish Dis. 41:11–26. [DOI] [PubMed] [Google Scholar]
- Gilmour A, Gogel B, Cullis BR, Thompson R. 2009. ASReml user guide release 3.0. Hemel Hempstead (UK): VSN International Ltd. [Google Scholar]
- Gonen S, Baranski M, Thorland I, Norris A, Grove H, Arnesen P, Bakke H, Lien S, Bishop SC, Houston RD. 2015. Mapping and validation of a major QTL affecting resistance to pancreas disease (salmonid alphavirus) in Atlantic salmon (Salmo salar). Heredity (Edinb). 115:405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimholt U, Larsen S, Nordmo R, Midtlyng P, Kjoeglum S, Storset A, Saebø S, Stet RJ. 2003. MHC polymorphism and disease resistance in Atlantic salmon (Salmo salar); facing pathogens with single expressed major histocompatibility class I and class II loci. Immunogenetics. 55:210–219. [DOI] [PubMed] [Google Scholar]
- Haugland O, Mikalsen AB, Nilsen P, Lindmo K, Thu BJ, Eliassen TM, Roos N, Rode M, Evensen O. 2011. Cardiomyopathy syndrome of atlantic salmon (Salmo salar L.) is caused by a double-stranded RNA virus of the Totiviridae family. J Virol. 85:5275–5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjeltnes B, Bang-Jensen B, Bornø G, Haukaas A, Walde SC. 2018. The Health Situation in Norwegian Aquaculture 2017. Norwegian Veterinary Institute 2018; Available from: https://www.vetinst.no/rapporter-og-publikasjoner/rapporter/2018/fish-health-report-2017 [Google Scholar]
- Houston RD, Haley CS, Hamilton A, Guy DR, Tinch AE, Taggart JB, McAndrew BJ, Bishop SC. 2008. Major quantitative trait loci affect resistance to infectious pancreatic necrosis in Atlantic salmon (Salmo salar). Genetics. 178:1109–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjøglum S, Larsen S, Bakke HG, Grimholt U. 2006. How specific MHC class I and class II combinations affect disease resistance against infectious salmon anaemia in Atlantic salmon (Salmo salar). Fish Shellfish Immunol. 21:431–441. [DOI] [PubMed] [Google Scholar]
- Kolawole AO, Sharma P, Yan R, Lewis KJ, Xu Z, Hostetler HA, Ashbourne Excoffon KJ. 2012. The PDZ1 and PDZ3 domains of MAGI-1 regulate the eight-exon isoform of the coxsackievirus and adenovirus receptor. J Virol. 86:9244–9254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Løvoll M, Wiik-Nielsen J, Grove S, Wiik-Nielsen CR, Kristoffersen AB, Faller R, Poppe T, Jung J, Pedamallu CS, Nederbragt AJ, et al. 2010. A novel totivirus and piscine reovirus (PRV) in Atlantic salmon (Salmo salar) with cardiomyopathy syndrome (CMS). Virol J. 7:309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moen T, Baranski M, Sonesson AK, Kjøglum S. 2009. Confirmation and fine-mapping of a major QTL for resistance to infectious pancreatic necrosis in Atlantic salmon (Salmo salar): population-level associations between markers and trait. BMC Genomics. 10:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ødegård J, Baranski M, Gjerde B, Gjedrem T. 2011. Methodology for genetic evaluation of disease resistance in aquaculture species: challenges and future prospects. Aquac Res. 42:103–114. [Google Scholar]
- Ødegård J, Meuwissen THE. 2012. Estimation of heritability from limited family data using genome-wide identity-by-descent sharing. Genet Sel Evol. 44:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegård J, Moen T, Santi N, Korsvoll SA, Kjøglum S, Meuwissen TH. 2014. Genomic prediction in an admixed population of Atlantic salmon (Salmo salar). Front Genet. 5:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ødegård J, Olesen I, Gjerde B, Klemetsdal G. 2007. Evaluation of statistical models for genetic analysis of challenge-test data on ISA resistance in Atlantic salmon (Salmo salar): prediction of progeny survival. Aquaculture. 266:70–76. [Google Scholar]
- Raymond B, Bouwman AC, Wientjes YCJ, Schrooten C, Houwing-Duistermaat J, Veerkamp RF. 2018. Genomic prediction for numerically small breeds, using models with pre-selected and differentially weighted markers. Genet Sel Evol. 50:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robledo D, Matika O, Hamilton A, Houston RD. 2018. Genome-wide association and genomic selection for resistance to amoebic gill disease in Atlantic Salmon. G3 (Bethesda). 8:1195–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodger HD, McCleary SJ, Ruane NM. 2014. Clinical cardiomyopathy syndrome in Atlantic salmon, Salmo salar L. J Fish Dis. 37:935–939. [DOI] [PubMed] [Google Scholar]
- Storset A, Strand C, Wetten M, Kjøglum S, Ramstad A. 2007. Response to selection for resistance against infectious pancreatic necrosis in Atlantic salmon (Salmo salar L.). Aquaculture. 272:62–68. [Google Scholar]
- Timmerhaus G, Krasnov A, Takle H, Afanasyev S, Nilsen P, Rode M, Jørgensen SM. 2012. Comparison of Atlantic salmon individuals with different outcomes of cardiomyopathy syndrome (CMS). BMC Genomics. 13:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HY, Hamilton A, Tinch AE, Guy DR, Bron JE, Taggart JB, Gharbi K, Stear M, Matika O, Pong-Wong R, et al. 2016. Genomic prediction of host resistance to sea lice in farmed Atlantic salmon populations. Genet Sel Evol. 48:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HY, Hamilton A, Tinch AE, Guy DR, Gharbi K, Stear MJ, Matika O, Bishop SC, Houston RD. 2015. Genome wide association and genomic prediction for growth traits in juvenile farmed Atlantic salmon using a high density SNP array. BMC Genomics. 16:969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsunomiya YT, do Carmo AS, Carvalheiro R, Neves HH, Matos MC, Zavarez LB, Pérez O’Brien AM, Sölkner J, McEwan JC, Cole JB, et al. 2013. Genome-wide association study for birth weight in Nellore cattle points to previously described orthologous genes affecting human and bovine height. BMC Genet. 14:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassgård S. 2017. Quantitative genetics of cardiac traits in Atlantic salmon (Salmo salar L.). Norwegian University of Life Sciences, Ås. Available from: https://nmbu.brage.unit.no/nmbu-xmlui/handle/11250/2453492 [Google Scholar]
- White E. 2001. Regulation of the cell cycle and apoptosis by the oncogenes of adenovirus. Oncogene. 20:7836–7846. [DOI] [PubMed] [Google Scholar]
- Wientjes YCJ, Bijma P, Vandenplas J, Calus MPL. 2017. Multi-population genomic relationships for estimating current genetic variances within and genetic correlations between populations. Genetics. 207:503–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez JM, Houston RD, Newman S. 2014. Genetics and genomics of disease resistance in salmonid species. Front Genet. 5:415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. 2011a. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 88:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Weedon MN, Purcell S, Lettre G, Estrada K, Willer CJ, Smith AV, Ingelsson E, O’Connell JR, Mangino M, et al. ; GIANT Consortium 2011b. Genomic inflation factors under polygenic inheritance. Eur J Hum Genet. 19:807–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used in this study were from a private breeding company, therefore we are not permitted to share phenotypic and genotypic data directly, however, data is available upon request from the corresponding author.