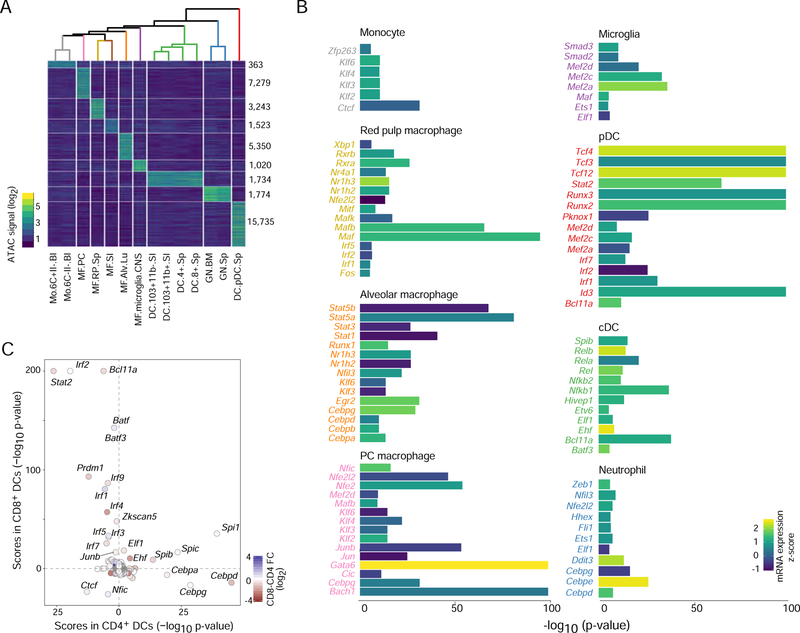
Figure 6. Regulatory factors in myeloid cells inferred from chromatin accessibility.
(A) Differential OCR signals across all steady-state myeloid cells (right: numbers of distinguishing OCRs; top: correlation tree). (B) TF motif enrichment scores (chromVAR z-test) in myeloid group-specific OCRs from A, filtered for TF expression levels and statistical significance, with signed -log10 p values capped at 100 for display; bars shaded by TF mRNA expression. (C) Comparison of TF enrichment scores for OCRs accessible CD4+ and CD8+ cDCs, points are shaded according to the TF’s mRNA expression fold change between the two DC subsets. See Fig. S6.