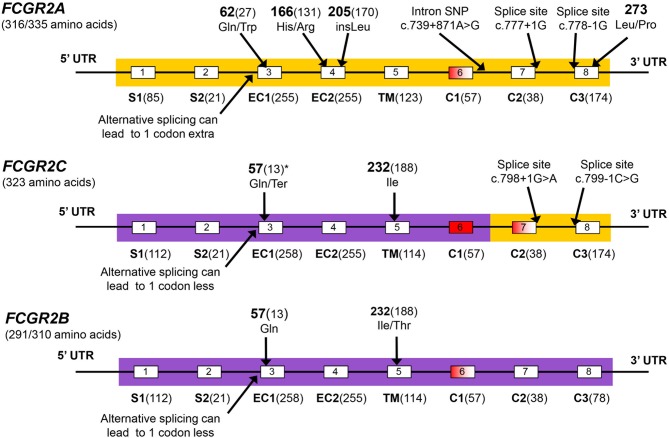
Figure 3.
FCGR2 exons. The FCGR2C gene is the crossover product from an unequal crossover between FCGR2A and FCGR2B. Coloring of FCGR2C matches the color of the other FCGR2 genes in the parts where it is highly homologous to that gene. Exons are shown by boxes, white exons are included in all transcripts, red exons are always spliced out, red-shaded exons are spliced out in some transcripts but retained in others. Exon names are below and followed by the number of coding base pairs in that exon. S1, S2, signal peptides; EC1, EC2, extracellular domains; TM, transmembrane domain; C1, C2, C3, cytoplasmic domains. The C3 exons contain an immunoreceptor tyrosine-based activation motif (ITAM) in FCGR2A and FCGR2C, and contains an immunoreceptor tyrosine-based inhibitory motif (ITIM) in FCGR2B. There is a potential confusion with regard to exon numbering in FCGR2 genes: In the FCGR2B gene, transcripts exist that do (FCGR2B1) or do not (FCGR2B2) retain the 57 bp exon6, dependent on the cell type in which the receptor is expressed. In FCGR2A and FCGR2C a very homologous exon6 is present on genomic level, but this exon is always spliced out in FCGR2A and FCGR2C transcripts. Only in some rare cases exon6 is retained in FCGR2A, which results in a gain-of-function FcγRIIA isoform (39). The final two exons of FCGR2A and -2C are often designated exon6 and 7, but to reflect the homology between the 3 FCGR2 genes, we chose to include the potential exon6 in the nomenclature of all FCGR2 genes, designating the final 2 exons exon7 and 8. However, we do not include the base pairs of this exon6 when indicating the nucleotide positions in the FCGR2A and FCGR2C transcripts, because this is not normally done in the literature. In FCGR2A transcripts, further inconsistencies exist as a result of alternative splicing at the beginning of exon3 because of two adjacent splice acceptor sites that can both be used. The most commonly used nucleotide and amino acid numbering is derived from the shorter transcript in which the 3' splice acceptor site is used, so we chose to use this transcript for nucleotide numbering throughout this thesis. Several SNPs are indicated in the figure, all are indicated by their amino-acid position in the full protein, followed by the amino acid position excluding signal peptides between brackets for some of the SNP which are commonly known by that position.
*The p.Gln57Ter SNP in FCGR2C is part of a haplotype of 8 SNPs in intron2 and exon3 in FCGR2C, this whole haplotype is identical to FCGR2B in the case of p.57Gln (40). p57Gln is the only non-synonymous coding SNP in this haplotype.