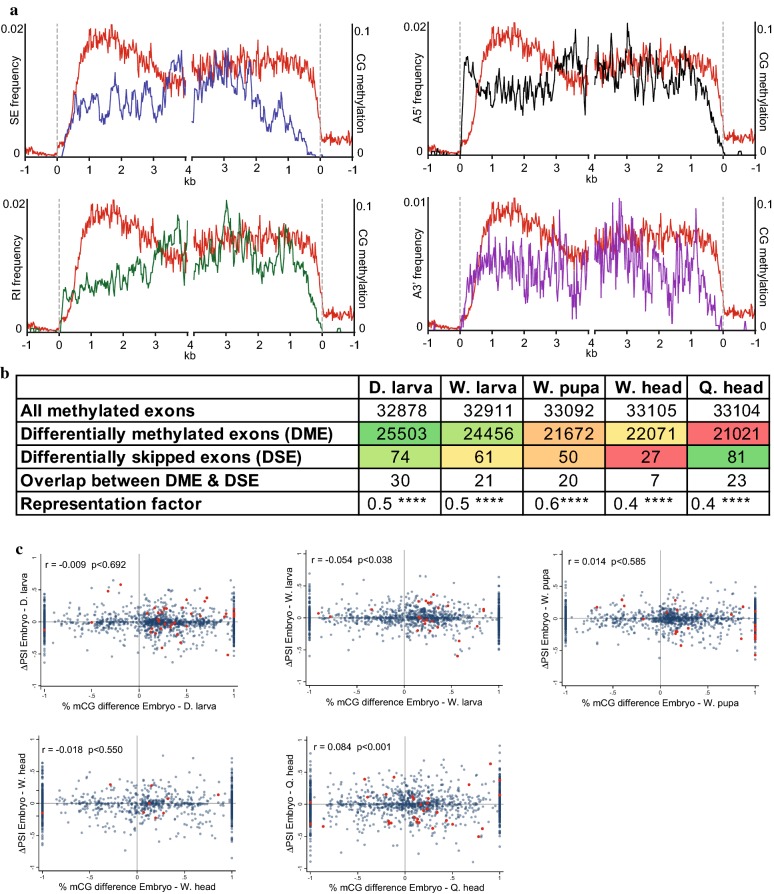
Fig. 5.
Gene body methylation dynamics are not associated with developmentally regulated RNA splicing patterns. a Skipped exons (blue), retained introns (green) and alternative 5′/3′ spliced sites (black/purple) frequencies were averaged in 20-bp bins along methylated genes (averaged genic methylation > 0.05) essentially as described in Fig. 1a. Averaged CG methylation profile is illustrated with red lines. b This table includes the number of all methylated exons (mCG > 0.05 in either embryo or the other biological sample (indicated on the top line of the columns), number of differentially methylated exons (DMEs; Fisher Exact Test p < 0.05 in both of biological replicates), number of differentially AS-skipped exons (DSEs), number of overlap exons between DMEs and DSEs, and representation factors, which is the ratio between the number of observed overlapped exons with that of the expected one. A representation factor < 1 means that it is under-representated. *Denote p value strength. c Scatter plots of delta PSI (ΔPSI) of exons between embryo and indicated samples versus percent-methylation-change of exons in the same pairwise samples. Blue dots include values of all methylated exons, and red dots include values of only differentially skipped and methylated exons (i.e., DSEs and DMEs)