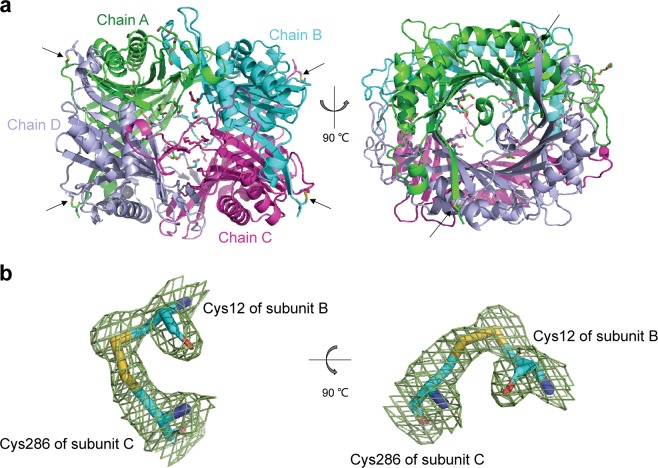
Fig. 5.
Structure of the K12C–E286C AgUricase mutant. The structure of the K12C–E286C AgUricase mutant was generated with the PyMOL program (PDB entry: 6OE8). a Side and top views of the schematic model of the AgUricase tetramer: chain A (TV green), chain B (cyan), chain C (purple), and chain D (light blue). Disulfide bonds are shown in yellow and indicated by black arrows. b Side and top views of the 2│Fo│−│Fc│density map of the disulfide bonds between Cys12 of subunit B and Cys286 of subunit C. Disulfide bonds are shown in yellow