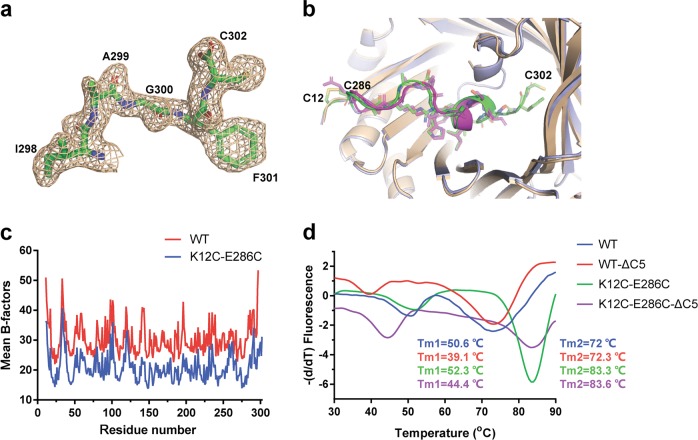
Fig. 6.
A C-terminal flexible loop is required for the stability of AgUricase protein. a The 2│Fo│−│Fc│density map of residues 298–302 in AgUricase. b Structural comparison of the C-terminal residues 286–302 in wild-type and K12C–E286 mutant AgUricase. Wild-type AgUricase is gray, and the K12C–E286C mutant AgUricase is light blue, and its C-terminal residues (286–302) are green and the disulfide bonds are yellow. c Mean B-factors of wild-type and K12C–E286C AgUricase. The mean B-factors of wild-type and K12C–E286C AgUricase were calculated according to the crystal structures of wild-type AgUricase (PDB ID 2YZB) and K12C–E286C AgUricase (PDB ID 6OE8). d Results of the thermal shift assay (TSA) of the △C5 deletion mutant of the K12C/E286C AgUricase compared with those of the K12C/E286C AgUricase