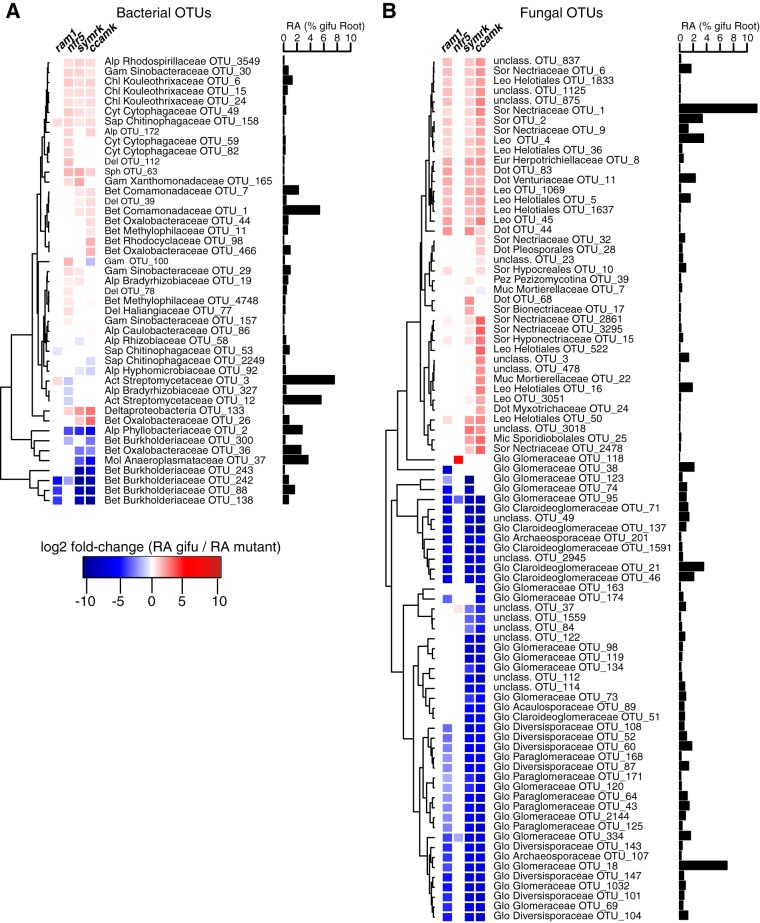
FIG 5.
Differential abundance analysis of root-associated OTUs. (A) Dendrogram of bacterial OTUs that are differentially abundant in the roots of mutants compared to WT roots. (B) Dendrogram of fungal OTUs that are differentially abundant in the roots of mutants compared to WT roots. Only OTUs that have an average RA of >0.1% across all root samples, including mutants, are considered here. The dendrogram is based on hierarchical clustering. For each OTU, the fold change in RA from the WT to mutants is indicated (P < 0.05 by a Kruskal-Wallis test). Next to each OTU, the RA in WT roots is indicated. Phylum and family associations (if available) are given for each OTU. Abbreviations of bacterial phyla: Del, Deltaproteobacteria; Gem, Gemm-1; Chl, Chloroflexi; Bet, Betaproteobacteria; Alp, Alphaproteobacteria; Gam, Gammaproteobacteria; Cyt, Cytophagia; Sap, Saprospiria; Ped, Pedosphaera; Sph, Sphingobacteria; Mol, Mollicutes. Abbreviations of fungal phyla: Sor, Sordariomycetes; Dot, Dothideomycetes; Mic, Microbotryomycetes; Ust, Ustilaginomycetes; Eur, Eurotiomycetes; Leo, Leotiomycetes; Aga, Agaricomycetes; Glo, Glomeromycetes; Pez, Pezizomycotina; Muc, Mucoromycotina. Data for ram1 AMS-deficient, nfr5 RNS-deficient, symrk AMS- and RNS-deficient, and ccamk AMS- and RNS-deficient plants are shown.