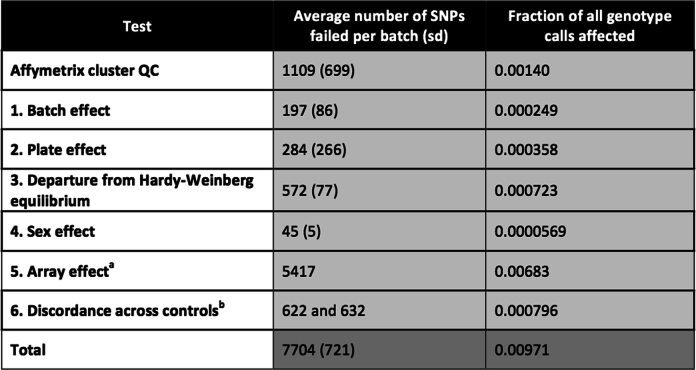
Extended Data Table 4.
Failure rates for six marker-based quality tests
For all numbered tests, a marker (or marker within a batch) was set to missing if the test yielded P < 10−12, except in the case of test 6, for which a marker was set to missing if the test yielded <95% concordance. See Supplementary Information for details of each test (n = 463,844 samples). The total is not equal to the sum of all tests because it is possible for a marker to fail more than one test. Because the two arrays contain slightly different sets of markers, the total number of genotype calls used to compute the fractions is:
Nukbb Lukbb + Nukbl Lukbl, in which N and L refer to the numbers of markers and samples typed on the UK Biobank Axiom array (ukbb) and samples typed on the UK BiLEVE Axiom array (ukbl) within the Affymetrix data delivery (see Supplementary Table 1).
aThe array effect test was applied across all batches and only for markers present on both arrays, so we simply report the total number of markers that failed this test.
bThe discordance test was applied across all batches, but not all markers are present on both arrays. The first value is the number of unique markers on the UK BiLEVE Axiom array that failed this test, and the second is for markers on the UK Biobank Axiom array.