Fig. 3.
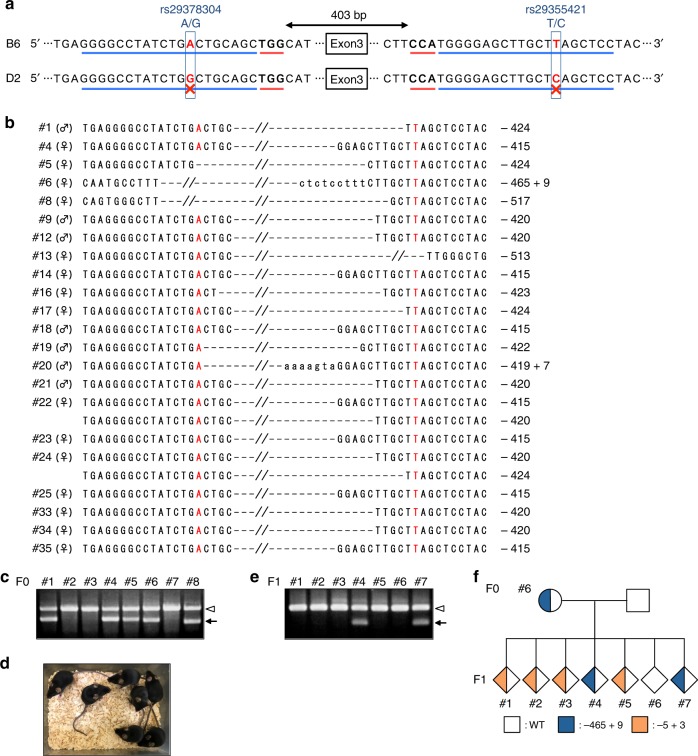
HypaCas9 enables efficient generation of monoallelic genome-modified mice. a Design of two gRNAs for B6 allele-specific deletion of Cdk1 exon 3. The blue line indicates the gRNA target site and the red line indicates the PAM of gRNA. SNP rs29378304 is located in intron 2 of Cdk1 and rs29355421 in intron 3. B6-targeting gRNAs were designed to have these SNPs within the protospacer. b Sequencing analysis of the deletion allele in F0 mice. The SNPs rs29378304 and rs29355421 are shown in red. c Representative PCR analysis of the Cdk1 exon 3 deletion in F0 mice. d Representative F0 mice including both mice with and without the deletion allele. e PCR analysis of the Cdk1 exon 3 deletion in F1 embryos, which were obtained by mating a #6 F0 female mouse with a WT male mouse. In c and e, the white arrowhead indicates the WT allele or an allele with a small indel. The arrow indicates the deletion allele. f Pedigree showing inheritance of the deletion allele of the #6 F0 mouse indicated in blue. Orange indicates the minor allele with a small indel (−5 + 3) caused by mosaic mutation, which is undetectable by sequencing in the #6 F0 mouse