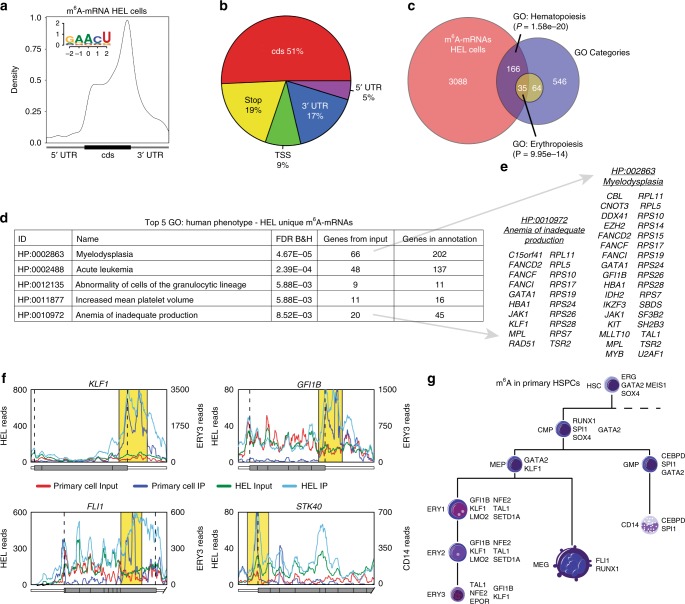
Fig. 2.
m6A marks the mRNA of key hematopoietic and erythroid regulators. a Distribution of m6A sites detected by meRIP-seq in HEL cells (150 µg), with an enrichment around the stop site, enriched m6A methylation site motif. b Pi chart displaying the frequency of m6A peaks, from HEL (150 µg), within different transcript regions: TSS, centered around translation start ATG; Stop, centered around the stop codon. c Genes uniquely methylated in HEL cells vs. 293T cells36,37 are enriched for key hematopoiesis and erythropoiesis genes. d Top gene ontology (GO) terms for HEL unique m6A mRNAs are enriched for genes with causal roles in hematopoietic diseases. e Gene callouts for GO terms identified in (d). f Transcript maps for erythropoiesis regulators detected as methylated in both HEL (150 µg) and adult BM cells show overlapping methylation patterns (yellow highlight). g Highlighting key hematopoietic regulators detected as methylated by meRIP-seq in flow sorted adult human BM cells. The parameters used to define the hematopoietic populations are outlined in Supplementary Fig. 2h