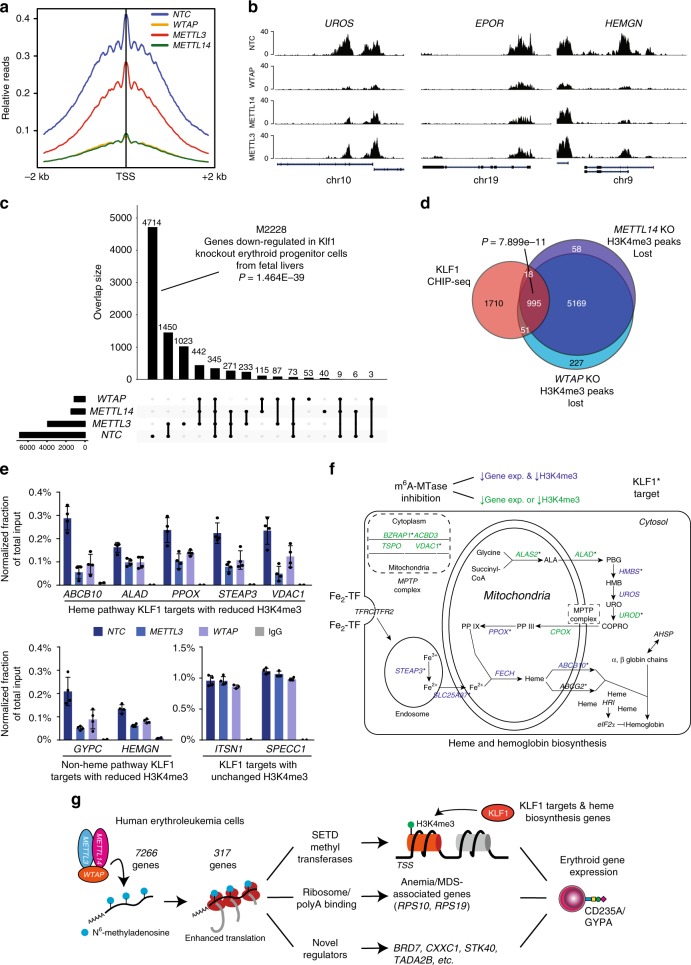
Fig. 5.
Promoter histone H3K4me3 marks are lost in the absence of m6A methyltransferase activity. a An H3K4me3 relative read distribution plot around the transcription start site (TSS) of methylated genes in CRISPR NTC, WTAP-KO, METTL3-KO, and METTL14-KO HEL cells. The plot shows a marked reduction in H3K4me3 marks following loss of WTAP and METTL14 with a more modest effect with loss of METTL3. b Bedgraphs of normalized H3K4me3 CUT&RUN data for select genes with reduced methylation following m6A loss. c An upset plot of the H3K4me3 peaks found in CRISPR NTC, WTAP-KO, METTL3-KO, and METTL14-KO HEL cells. The pattern of H3K4me3 loss is consistent with loss of KLF1 transcriptional regulation. Genes with reduced H3K4me3 following loss of any of the three m6A MTase components are enriched for genes downregulated following Klf1 KO. d A Venn diagram showing enrichment for KLF1 CHIP-seq target genes63 among the H3K4me3 peaks lost following WTAP-KO or METTL14-KO, suggesting m6A-mediated epigenetic regulation of the KLF1 transcriptional program. e KLF1 CHIP-qPCR validation of several genes with or without altered H3K4me3 levels following METTL3-KO and WTAP-KO, (n = 4, mean ± SEM). f A diagram of the iron procurement, heme synthesis and transport, and hemoglobin assembly in erythroid cells, highlighting regulation by KLF1 and altered H3K4me3 marking following m6A loss. *: KLF1 CHIP-seq target genes63; Blue: reduced H3K4me3 marking and transcript expression following m6A-MTase inhibition; Green: reduced H3K4me3 marking or transcript expression following m6A-MTase inhibition. These results highlight inhibition of multiple pathways involved in hemoglobin synthesis following loss of m6A possibly through down regulation of the KLF1 transcriptional program. g The proposed model for the role of m6A in translational regulation of erythroid gene expression and erythropoiesis