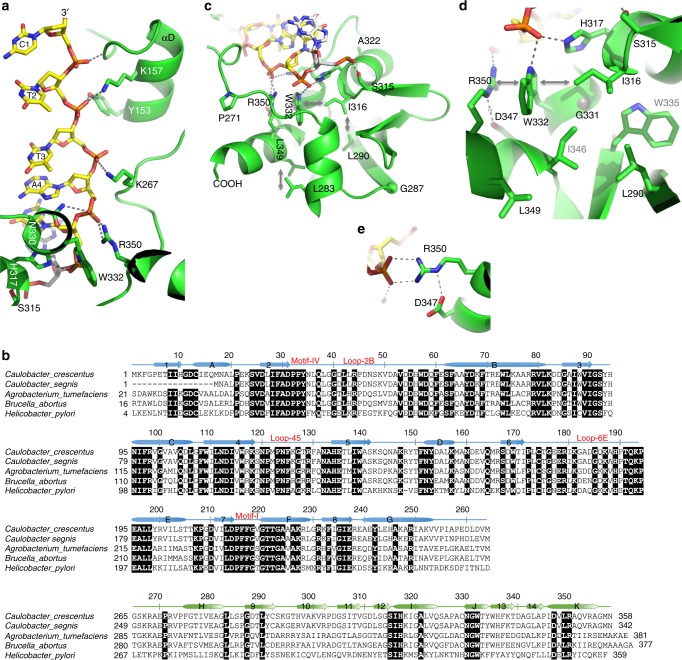
Fig. 5.
Sequence conservation of CcrM orthologs. a Molecule B (helix αD and C-terminal domain) provides almost all DNA phosphate interactions along the non-target strand. b Sequence alignment of five CcrM orthologs: Caulobacter crescentus (NC_002696), Caulobacter segnis (NC_014100.1), Agrobacterium tumefaciens (NC_003062), Brucella abortus (AF011895), and Helicobacter pylori (AGR62562). Invariant residues are marked with white letters on black background. c-e Structural features of invariant residues in the C-terminal domain. c Pro271 and Gly287 are structural residues located in the linker loop and the beginning of strand β9. d Leu283, Leu290, Ile316, Gly331, Trp332, and Leu349 are part of the hydrophobic core that supports the integrity of the C-terminal domain. e Asp347 and Arg350 form an electrostatic interaction