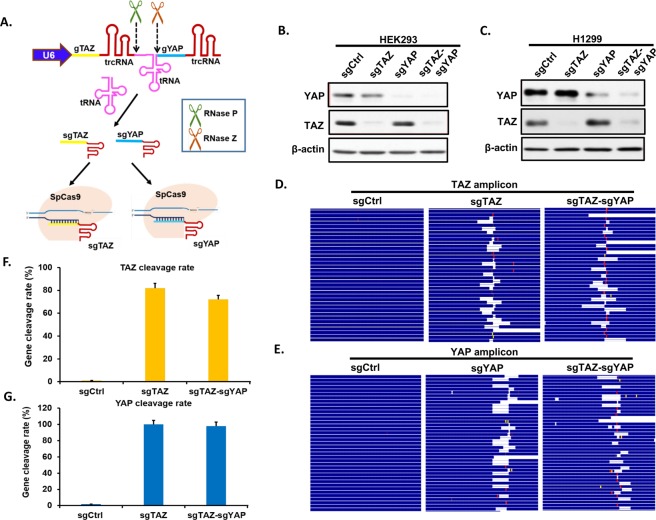
Figure 1.
Efficient tRNA-gRNA CRISPR system in mammalian cell lines. (A) A schematic showing how tRNA-gRNA system works in mammalian cells. sgRNAs targeting TAZ (sgTAZ) and YAP (sgYAP) are linked through tRNA to form a single transcript driven by U6 promoter. The tRNA precursor is cut off from the transcript through the endogenous RNase P and RNase Z, during which sgTAZ and sgYAP are separated to form complexes with Cas9 individually to target relative genes with complimentary sequences. See also Fig. S1. (B,C) HEK293 (B) and H1299 (C) cells were infected with lentiviral constructs expressing a single sgRNA targeting individual genes (sgTAZ, sgYAP) or a tRNA franked with two gRNAs targeting both TAZ and YAP (sgTAZ-sgYAP). Cell lysates were extracted after 14 days post infection/selection and subjected to western blotting for detection of YAP and TAZ protein expression levels compared with lysates from sgControl (Ctrl) infected cells. β-actin was used as loading control. (D–G) Genomic DNAs (gDNAs) were extracted from HEK293 infected with lentiviral constructs of single sgTAZ, sgYAP, and double sgTAZ-sgYAP after 14 days culture post infection/selection. TAZ (D,F) and YAP (E,G) amplicons containing relative sgRNA targeting regions were amplified and sent for next generation sequencing (NGS). Sequences of relative genes were further mapped to relative gene locus to check the efficiency of both single and double CRISPR system (D,E). The blank indicates deletions, red bars indicate mutations and yellow bars represent insertions. The cleavage of both TAZ (F) and YAP (G) genes were counted and cleavage rate was calculated and presented in the charts.