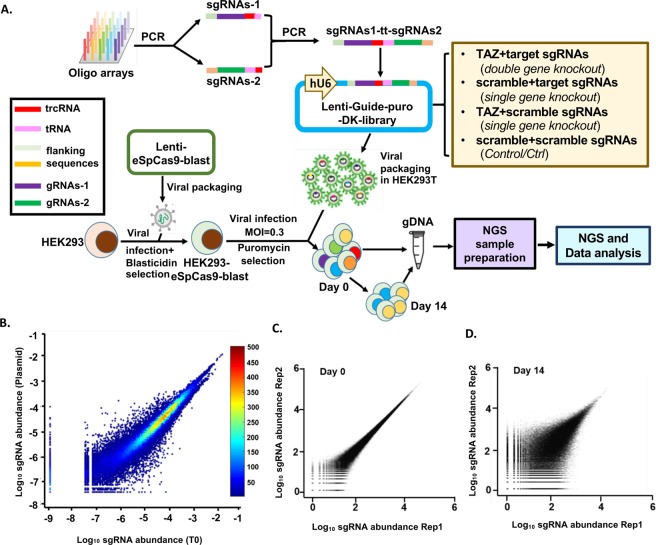
Figure 2.
Establishment and evaluation of dual gRNA library targeting TAZ-genome through TCGI method. (A) A schematic method for establishing TCGI platform for differential growth screen. Array-synthesized oligos were amplified through PCR with relative primers. Then two types of PCR pools were assembled together into full length of segments for cloning into pLenti-Guide vector to generate the lenti-Guide-puro-double knockout (DK) library, including both double gene knockout and single gene knockout as clarified in the figure. HEK293 cells were transduced with lentivirus plasmid containing eSpCas9 and blasticidin resistant gene to generate HEK293-eSpCas9-blast cell line, which stably expresses eSpCas9. Then, the DK library was transduced into HEK293-eSpCas9 cells. After puromycin selection (Day 0), these cells were further cultured for 14 days (Day 14) and gDNA was extracted from both Day0 as well as Day 14 for NGS sample preparation. Deep sequencing data from NGS were analyzed to find out genes synergistically affect cell growth. (B) Correlations of double-sgRNA constructs in the DNA library (Plasmid) and after transduction and puromycin selection (T0). Spearman Correlation = 0.977. Color bar shows the density of data. (C,D) Characterization of biological replicates of the double-sgRNA libraries (Rep1 and Rep2, two biological replicates) at day 0 (C) and at day 14 (D).