Abstract
Pathogens often receive antibiotic resistance genes through horizontal gene transfer from bacteria that produce natural antibiotics. ErmE is a methyltransferase (MTase) from Saccharopolyspora erythraea that dimethylates A2058 in 23S rRNA using S-adenosyl methionine (SAM) as methyl donor, protecting the ribosomes from macrolide binding. To gain insights into the mechanism of macrolide resistance, the crystal structure of ErmE was determined to 1.75 Å resolution. ErmE consists of an N-terminal Rossmann-like α/ß catalytic domain and a C-terminal helical domain. Comparison with ErmC’ that despite only 24% sequence identity has the same function, reveals highly similar catalytic domains. Accordingly, superposition with the catalytic domain of ErmC’ in complex with SAM suggests that the cofactor binding site is conserved. The two structures mainly differ in the C-terminal domain, which in ErmE contains a longer loop harboring an additional 310 helix that interacts with the catalytic domain to stabilize the tertiary structure. Notably, ErmE also differs from ErmC’ by having long disordered extensions at its N- and C-termini. A C-terminal disordered region rich in arginine and glycine is also a present in two other MTases, PikR1 and PikR2, which share about 30% sequence identity with ErmE and methylate the same nucleotide in 23S rRNA.
Subject terms: Enzymes, Antimicrobial resistance, X-ray crystallography
Introduction
The ribosome is the large macromolecular machine responsible for the sequential template-dependent polymerization of amino acids into a polypeptide chain1, and an important drug target for antibiotics2,3. Macrolide antibiotics and their third-generation ketolide derivatives are used against a broad range of Gram-positive pathogens. They inhibit translation by binding in the nascent peptide exit tunnel close to the peptidyl transferase center of the large ribosomal subunit4,5. One important mechanism of microbial resistance to macrolides is the N6 methylation of 23S rRNA nucleotide A2058 (Escherichia coli numbering) in the macrolide binding site by the Erm (erythromycin ribosome methylation) group of MTases5,6. This group of enzymes uses SAM to specifically mono- or dimethylate a 50S ribosomal precursor substrate7,8, where A2058 is accessible for modification. Erm genes were originally identified in microorganisms producing natural macrolides as a mechanism of self-protection against their own antibiotics9. ErmE (EC 2.1.1.184) is a dimethyltransferase from the actinomycete Saccharopolyspora erythraea, from which the first macrolide antibiotic erythromycin was originally extracted10,11. ErmE provides resistance to macrolide, lincosamide, and streptogramin B (MLS) antibiotics to S. erythraea12. However, in conditions of wide use of antibiotics, horizontal gene transfer has led to propagation of pathogens carrying this and other erm genes.
Ketolides were initially developed to overcome macrolide resistance13 and present a very promising class of antibiotics14. Most members of this class are synthetic and semi-synthetic derivatives of macrolides. However, Streptomyces venezuelae strain ATCC 15439 produces the natural ketolide pikromycin15. To avoid self-inhibition, this microorganism expresses two MTases PikR1 and PikR2 that mono- and dimethylate A205816, the same nucleotide as ErmE. PikR1 and PikR2 display 39% sequence identity to each other and 35% and 33% sequence identity to ErmE.
In light of the danger of horizontal transfer of macrolide and ketolide resistance genes, there is an urgent need for better understanding of the respective resistance mechanisms, including information on the structural and functional properties of ribosome-modifying enzymes. Here, we present the crystal structure of rRNA MTase ErmE, and analyze the similarities and differences to PikR1, PikR2, ErmC’ and other similar MTases.
Materials and Methods
DNA constructs, protein expression and purification
All codon-optimised N-terminally His8-tagged constructs were synthesized by GenScript and subcloned into the pET-24a(+) vector (Supplementary Table S1).
Plasmids were transformed into E. coli BL21(AI). Cultures were grown at 37 °C in LB media with 0.025 mg/ml kanamycin and 0.1% (w/v) D-glucose until an OD600 of 0.6. Protein expression was induced with 0.1% (w/v) L-arabinose. After overnight cultivation at 18 °C, the cells were collected by centrifugation, resuspended in lysis buffer (50 mM phosphate buffer pH 8, 1 M NaCl and 2 mM ß-mercaptoethanol) supplemented with 10 mM imidazole, 10% (v/v) glycerol, 0.06 mg/ml DNAse and cOmplete protease inhibitor cocktail (Roche, Switzerland), and lysed in a high-pressure homogenizer (Constant System Ltd, UK). The lysate was centrifuged for 1 h at 30,000 g and the supernatant was applied to a gravity-flow column containing Ni-sepharose resin (GE Healthcare, Sweden) equilibrated with lysis buffer and 10% (v/v) glycerol. The column was washed with lysis buffer containing 20 and 30 mM imidazole and protein elution was performed with 500 mM imidazole in 50 mM phosphate buffer pH 8, 0.3 M NaCl and 2 mM ß-mercaptoethanol. Eluted protein was dialysed against 20 mM Tris-SO4 pH 8, 0.8 M (NH4)2SO4, 2 mM ß-mercaptoethanol and loaded on a 5 ml HiTrap Phenyl HP column (GE Healthcare, Sweden) equilibrated with dialysis buffer. Elution was done with a linear gradient of (NH4)2SO4 (0.8–0 mM) in 20 mM Tris-SO4 pH 8. Size-exclusion chromatography (SEC) was performed using a HiLoad 16/600 Superdex 75 pg column (GE Healthcare, Sweden) equilibrated with running buffer (20 mM Tris-SO4 pH 8, 0.3 M (NH4)2SO4 and 2 mM ß-mercaptoethanol). Peak fractions were analysed with SDS-PAGE and concentrated to 10 mg/ml using a 10 kDa cutoff Vivaspin Turbo concentrator (Sartorius, Germany). Purification was performed at 4 °C.
Differential scanning fluorimetry (DSF)17 was done using a BioRad CFx connect real time PCR machine.
Crystallization, data collection and structure determination
All proteins were subjected to sitting drop vapor diffusion crystallization using a mosquito crystallization robot (TTP Labtech, UK). Rhomboid-shaped tetragonal crystals of truncated ErmE grew in 5 d at room temperature in drops of 200 nl in 2% (v/v) tacsimate pH 5.0, 0.1 M sodium citrate tribasic dihydrate pH 5.6 and 16% (w/v) PEG 3350 (PEG/Ion screen, Hampton Research, US). For data collection at beamline ID30A-3 (MASSIF-3)18 of the European Synchrotron Radiation Facility (Grenoble, France), the crystal was fished directly from the drop and flash frozen in liquid nitrogen. X-ray experiments were done at 0.9677 Å wavelength at 100 K.
Data was processed using XDS19. The structure was solved by molecular replacement with Phaser20, using as search model an ensemble generated from PDB IDs 1QAM21, 3FUU22, 1YUB23, 3FYC24 and 1ZQ9 by CCP4 online pipeline MrBump25. The structure was traced with PHENIX AutoBuild26 followed by completion of missing regions in ARP/wARP27. Manual rebuilding was done in Coot28 and refinement with phenix.refine29. Protein geometry was validated in MolProbity30. All figures representing structures were made using PyMOL31.
Data collection and refinement statistics are reported in Table 1. A stereo image of a section of the 2mFo-Dfc map is presented in Supplementary Fig. S1.
Table 1.
Data collection and refinement statistics. Values in parentheses are for highest-resolution shell.
| Data collection | |
|---|---|
| No. of crystals | 1 |
| Space group | P43212 |
| Cell dimensions | |
| a, b, c; Å | 76.04, 76.04, 104.92 |
| α, β, γ; ° | 90, 90, 90 |
| Resolution, Å | 37.55–1.75 (1.81–1.75) |
| Rmerge‡ | 0.093 (3.79) |
| Rpim§ | 0.025 (0.98) |
| I/σI¶ | 19.04 (0.77) |
| Wilson B factor, Å2 | 34.15 |
| Total reflections | 474,706 (48,958) |
| Unique reflections | 31,708 (3,116) |
| Completeness, % | 99.91 (99.84) |
| Redundancy | 15.0 (15.7) |
| CC(1/2)# | 1 (0.324) |
| Refinement | |
| Resolution, Å | 37.55–1.75 |
| Reflections | 31,695 (3,112) |
| Free reflections | 1,829 (180) |
| Rworkll/Rfree** | 0.1920/0.2185 |
| Ramachandran plot | |
| Favored, % | 98.76 |
| Allowed, % | 1.24 |
| Outliers, % | 0.00 |
| No. of atoms | |
| Protein | 2,039 |
| Ligand | 5 |
| Water | 212 |
| B-factors | |
| Protein | 42.87 |
| Ligand | 84.84 |
| Water | 42.37 |
| R.m.s deviations | |
| Bond lengths, Å | 0.004 |
| Bond lengths, Å | 0.59 |
| PDB ID code | 6NVM |
‡Rmerge, ΣhklΣi|Ii(hkl) − 〈I(hkl)〉|/ΣhklΣi Ii(hkl), where Ii(hkl) is the intensity for an observation of a reflection and 〈I(hkl)〉 is the average intensity of all symmetry-related observations of a reflection.
§Rpim, Σhkl √(1/n − 1) Σi|Ii(hkl) − 〈I(hkl)〉|/ΣhklΣi Ii(hkl).
¶I/σI, signal to noise ratio.
#CC(1/2), percentage of correlation between intensities from random half-datasets.
llRwork, Σhkl||Fobs| − k|Fcalc||/Σhkl|Fobs|.
**Rfree, as Rwork, calculated from the free reflections excluded from refinement.
Results and Discussion
PikR1, PikR2 and ErmE purification and analyses
After initial purification tests, DSF measurements showed that the thermal stability of PikR1 increased in presence of phosphate and sulphate. For this reason, phosphate buffer was used during lysis and Tris-SO4 was used after IMAC, to avoid formation of salt crystals during crystallization.
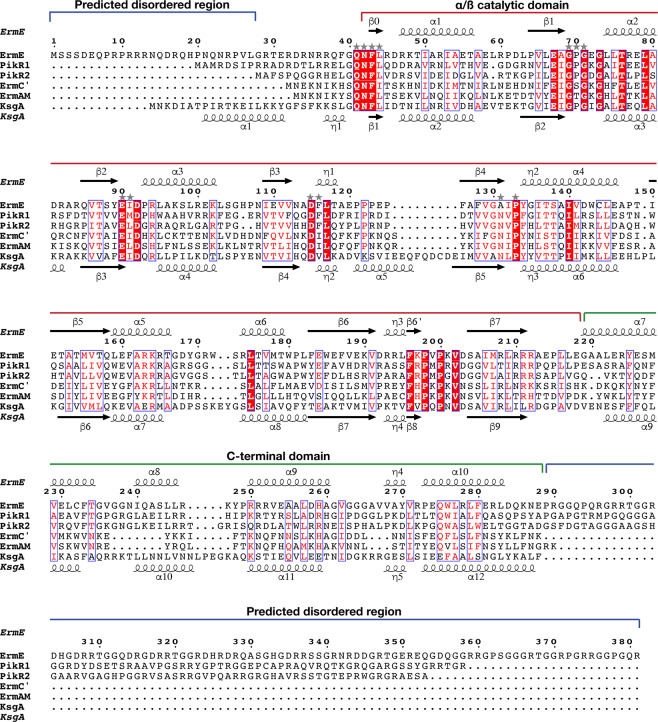
Full-length PikR1 purified using IMAC and HIC was analysed by SDS-PAGE and reproducibly showed two distinct bands (Fig. 1a). To determine the content of the bands and to exclude the presence of another protein, the two bands were subjected to mass-spectrometry analysis at the Proteomics Core Facility at University of Gothenburg (Sweden). The results demonstrated that both bands consisted of PikR1. Since the second, smaller, band was present after IMAC purification of the N-terminally His8-tagged PikR1, we hypothesized that it was the result of a C-terminal proteolytic degradation. In support of this, investigation of the PikR1 sequence with the PrDOS online server32 predicted disorder of a C-terminal region of around 67 aa, which could make the protein susceptible to proteolytic degradation as well as prevent crystallization of the full-length protein. For PikR2 and ErmE, C-terminal regions of 64 and 93 residues were similarly predicted to be disordered (Fig. 2).
Figure 1.
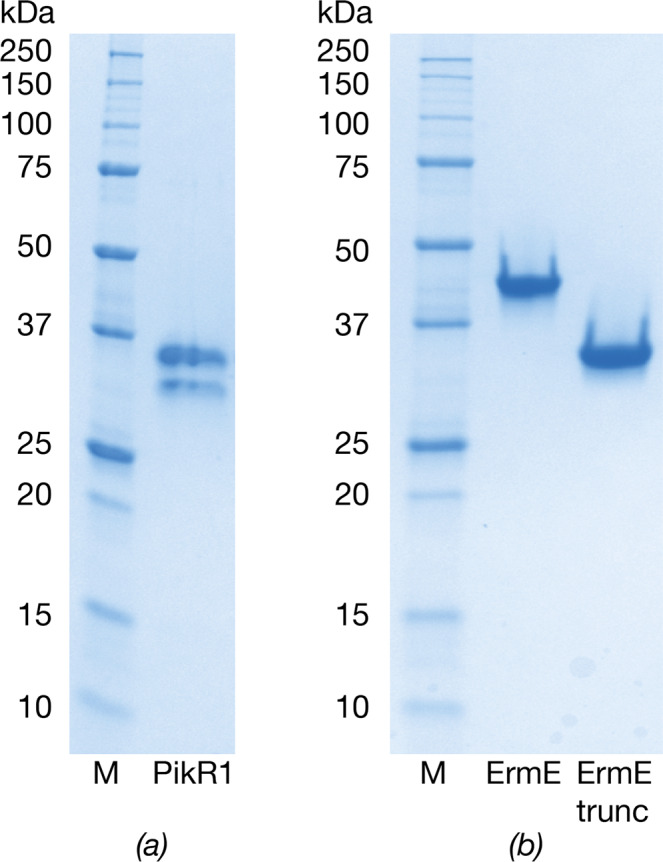
Coomassie-stained SDS-PAGE. (a) PikR1 after HIC purification. (b) Full-length and truncated ErmE after SEC purification. M: Precision Plus Dual Color Standard (BioRad). Full-length gels are presented in Supplementary Fig. S2.
Figure 2.
Structure-guided sequence alignment of ErmE with PikR1, PikR2, ErmC’ (PDB 1QAM)21, ErmAM (PDB 1YUB)23 and KsgA from B. subtilis (PDB 6IFT)52. Domain organization and secondary structure elements of ErmE are shown above the alignment and secondary structure of KsgA below the alignment. Conserved residues are highlighted with white text on red background and conservative substitutions are presented by red text on white background. Residues predicted to interact with SAM are indicated with stars. The alignment was done with Expresso53 and visualized with ESPript 3.054. Disordered regions of ErmE were predicted with PrDOS32.
Based on this observation, full-length and C-terminally truncated variants of PikR1, PikR2 and ErmE were expressed, purified and subjected to crystallization experiments. Only the truncated version of ErmE (Fig. 1b) produced diffraction-quality crystals.
Overall structure of ErmE
Crystals of the N-terminally His8-tagged construct of C-terminally truncated ErmE (UniProt ID P07287) including residues 1–290 (Supplementary Table S1) diffracted to 1.75 Å and belonged to space group P43212 with one molecule per asymmetric unit. The structure was solved by molecular replacement using an ensemble of structures with rRNA N6A-methylating activities. The refined structure includes residues 42–285 of ErmE. The absence of ordered density for the N- and C-termini confirms the predicted flexibility of these regions. SEC analysis and examination of the structure in PDBe PISA33 confirm that ErmE is a monomer.
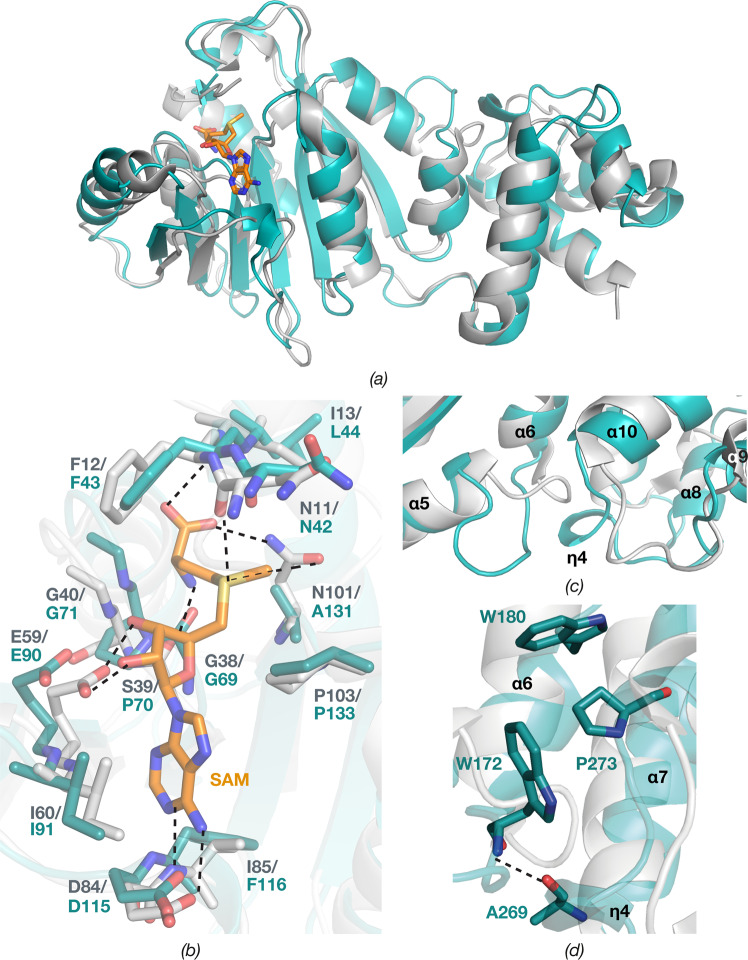
The bilobed structure of ErmE consists of an N-terminal Rossmann-like α/ß catalytic domain (residues 42–211) and C-terminal helical domain (residues 219–285), which are connected by a loop (Fig. 3). DALI34 identified dimethyltransferase ErmC’ (EC 2.1.48) from Bacillus subtilis (PDB ID 1QAM)21 as the structure most similar to ErmE, with root mean square deviation (rmsd) of 2.75 Å over 230 Cα atoms of the superposed structures (Fig. 4a). Interestingly, ErmE and ErmC’ share only 24% sequence identity despite having the same function and modifying the same site in 23S RNA. Other similar structures identified by DALI are 16S rRNA A1518 and A1519 MTase KsgA (PDB ID 3FUV, rmsd 2.42 Å over 216 Cα atoms)22 and its archaeal homologue Dim1 (PDB ID 3FYC, rmsd 2.70 Å over 219 Cα atoms)24. Since these enzymes modify a different RNA substrate, we decided to mainly compare the ErmE structure to the structure of ErmC’.
Figure 3.
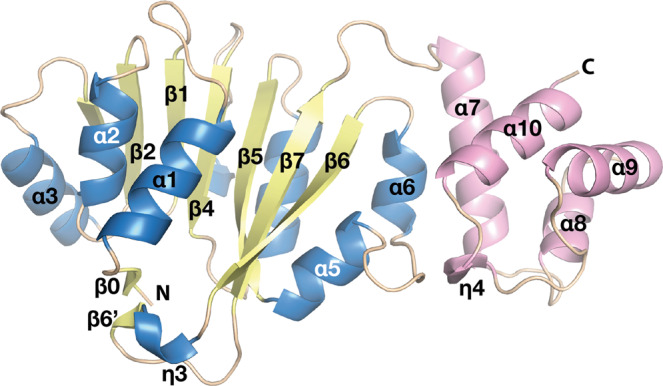
Cartoon representation of overall structure of ErmE. α-helices and ß-strands of the N-terminal α/ß catalytic domain are colored in blue and yellow; α-helices of the C-terminal domain are in pink and loops in wheat.
Figure 4.
Superposition of ErmC’ (grey) in complex with SAM (orange) (PDB ID 1QAO)21 onto ErmE (teal). (a) Overall structures. (b) Residues interacting with SAM at the SAM binding site. Main chain carbonyl oxygens are only shown if involved in SAM binding. Hydrogen bonds are shown by dashed lines. (c) Comparison of interdomain region including the α5-α6 and α9-α10 loops and the additional η4 in ErmE. (d) Residues involved in interdomain interactions in ErmE.
The Rossmann-like fold is common for nucleotide-binding proteins in general35, and the most common fold of the catalytic domain of SAM-dependent MTases36. The C-terminal domain was in ErmC’ proposed as an RNA-binding domain37 based on its large positively charged surface. However, it was later shown by mutagenesis that the key residues for specific RNA binding are located in the catalytic domain, facing the cleft between the domains. Accordingly, the C-terminal domain was suggested to mainly function in structural stabilization of the catalytic domain38.
N-terminal catalytic domain
The catalytic domain consists of seven parallel (ß1-ß6 and ß6’) and two antiparallel (ß0 and ß7) ß-strands that are surrounded by three α-helices (α1-α3) and one 310 helix (η3) on one side, and three α-helices (α4-α6) and two 310 helices (η1-η2) on the opposite side (Figs 2 and 3).
ErmE and ErmC’ have the same topology of their catalytic domains (Fig. 4a) that share 26% sequence identity. The domains superpose with rmsd of 1.65 Å over 155 Cα atoms, the main difference being a longer loop between helices α5 and α6 in ErmE.
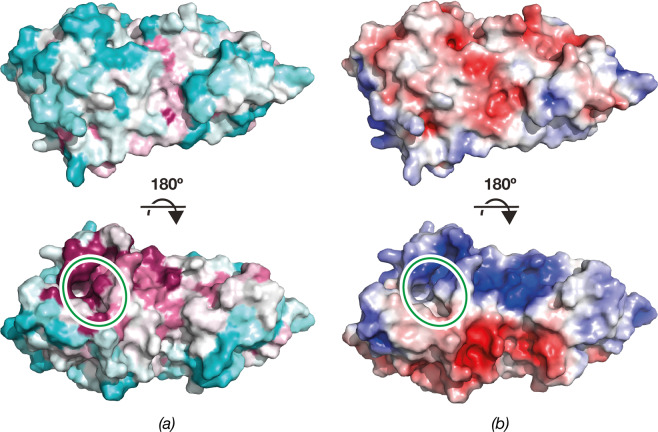
The N-terminal domain shows an L-shaped pocket rich in conserved residues (Figs 4a and 5). Conserved areas containing positively charged residues are also found above and to the right of the pocket (Fig. 5a,b), suggesting that these regions are involved in binding of the rRNA substrate.
Figure 5.
Surface representation of ErmE. (a) ErmE surface colored according to sequence conservation calculated by ConSurf55. The color spectrum ranges from magenta (highest conservation) to cyan (lowest conservation). (b) ErmE surface colored by electrostatic potential. The color spectrum ranges from deep red (−5 kT) to deep blue (+5 kT). The SAM binding site is indicated by a green ellipse.
SAM binding site
Superposition of the catalytic domains of ErmE and ErmC’ in complex with SAM suggests that SAM will bind similarly in the lower part of the pocket, with the methyl group directed towards the upper part of the pocket (Fig. 4a,b), where the substrate adenine will likely bind. Analysis of the ErmE structure with the 3DLigandSite server39 predicts the same binding site for SAM. Most SAM-binding residues are conserved from ErmC’, with minor differences presented by Ile13/Leu44 and Ser39/Pro70, involved in main-chain interactions with SAM, and Ile85/Phe116, where the side chain makes a hydrophobic interaction with the adenine group of SAM (Fig. 4b). Interestingly, the substitution of Asn101 in ErmC’ with Ala131 in ErmE suggests that the carboxyl group of SAM will only be coordinated through a hydrogen bond to the main chain N of Leu44, while in ErmC’ the same carboxyl in addition hydrogen bond to Asn101 Nδ2.
These residues are part of the sequence motif IV40 131AIPY134 in ErmE and 101NIPY104 in ErmC’ (Fig. 2), that is observed for example in dimethylating RNA or DNA N6-MTases (consensus sequence (A/S/N)(L/I/V)P(Y/F)41). Intriguingly, PikR1 that was reported to be a monomethylating MTase16, instead of the (N/D)PP(Y/F/W) motif associated with monomethylating N6A-MTases, contains the same NVPF motif as the dimethyltransferase PikR2 (Fig. 2) and both proteins are assigned to the same Pfam42 family of RNA dimethylases (PF00398).
C-terminal domain
In ErmE, the C-terminal domain is built from four α-helices (α7-α10) and one 310 helix (η4) (Figs 2 and 3). The C-terminal domains of ErmE and ErmC’ have similar topology, and despite only 18% sequence identity superpose with a rmsd of 1.78 Å over 53 Cα atoms. Thus, the longer α8 in ErmE corresponds to the short η4 in ErmC’. Interestingly, ErmC’ has a deletion at the position of the FTG tripeptide in α7 of ErmE that is conserved in both PikR MTases (Fig. 2). Another feature of ErmE is a longer loop with an inserted η4 helix between α9 and α10 (Fig. 3), where ErmC’ only contains a shorter loop (Fig. 4c).
Together with the C-terminal loop, η4 participates in interactions with the loop between α5 and α6 of the catalytic domain, contributing to stabilising the structure of ErmE (Fig. 3). A hydrogen bond is formed between main chain atoms of Ala269 and Trp172 and a hydrophobic interaction between Pro273, Trp172 and Trp180 (Fig. 4d). In ErmC’, the corresponding interaction is different due to the absence of η4 (Fig. 4c), and the involved residues are not conserved (Fig. 2).
In addition, the difference in interactions between N- and C-terminal domains in ErmE and ErmC’ leads to the slightly different orientation of these domains relative to each other, which results in a higher rmsd value for the superposition of the whole MTase structures as compared to when the individual domains are superposed.
Recognition of substrate RNA
In addition to ErmE, ErmC’ and ErmAM that provide antibiotic resistance, structures are available of two bacterial rRNA N6A-MTases involved in ribosome biogenesis, KsgA43 and RlmJ44 and catalytic domains of the human mRNA N6A-MTases METTL3-METTL1445 and METTL1646–48. These enzymes all display similar structures of their catalytic domains but make use of a variety of loops, tails or extra domains for specific recognition of the sequence and structure of their respective RNA substrates. There are few structures of N6A MTases in complex with RNA; KsgA49,50 and METTL1647. KsgA methylates a close to mature 30S subunit and similarly to the Erm family of MTases contains a C-terminal helical domain. For ErmE, it has been shown that although the natural substrate is a precursor of the 50S ribosomal subunit, the enzyme can specifically methylate a 27-nucleotide stem loop RNA substrate mimicking the local environment of A205851. Thus, some essential recognition elements in the RNA are located in close proximity to the adenosine that is methylated.
On the protein side, mutational studies on ErmC’ showed that a single arginine in equivalent position to Lys164 in α5 of ErmE is essential for erythromycin resistance38. In the ErmE structure, a sulfate ion is bound between Lys164 and Arg174, possibly mimicking a substrate phosphate. The positively charged surface of the C-terminal domain is also likely to contribute to substrate binding.
The N-terminal disordered region of ErmE is rich in arginine, while the C-terminal disordered region is dominated by glycine and arginine. Predicted disordered low-complexity regions with similar characteristics are also present in PikR1 and PikR2. These regions may contribute to binding of the 50S ribosome assembly intermediate where A2058 is accessible for modification. Similarly to the positively charged tails of ribosomal proteins, they may order upon interaction with the negatively charged RNA backbone. However, ErmC’ does not contain the corresponding long tails but can still recognize and modify the same substrate (Fig. 2).
Recently, KsgA was engineered to alter its substrate specificity and allow activity on the Erm substrate52. The strategy was based on exchanging the C-terminal domain, the N-terminal tail including α1 and η1 and the loop between α7 and α8 in KsgA to the corresponding sequences from ErmC’. The structure-guided sequence alignment of ErmE with ErmC’, ErmAM and KsgA (Fig. 2) shows that the Erm family enzymes, despite methylating the same substrate, display large variation in sequence and length in the N-terminus and the α7- α8 loop (KsgA numbering). This suggests that, out of the exchanged regions, the C-terminal domain is the more characteristic sequence element for the MTases that display specificity for each RNA substrate.
Conclusions
Here, we present the first crystal structure of rRNA methyltransferase ErmE, determined at 1.75-Å resolution. The structure of the enzyme could be potentially used for structure-based drug design with the aim to prevent macrolide antibiotic resistance in pathogens. Considering its higher than 30% sequence identity to PikR1 and PikR2, the structure of ErmE is also expected to be useful as a molecular replacement search model for further studies of PikR MTases.
Supplementary information
Acknowledgements
This work was supported by grant 2017-03827 and 2016-06264 from the Swedish Research Council to MS. We acknowledge the European Synchrotron Radiation Facility for access to beamline ID30A-3 and Sandesh Kanchugal and Daniel Larsson for constructive comments on the manuscript. Open access funding provided by Uppsala University.
Author Contributions
A.S. and M.S. designed the project. A.S. planned and carried out the experiments. A.S. and M.S. wrote the manuscript.
Data Availability
Atomic coordinates of ErmE have been deposited in the Protein Data Bank with accession code 6NVM.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Alena Stsiapanava, Email: alena.stsiapanava@gmail.com.
Maria Selmer, Email: maria.selmer@icm.uu.se.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-51174-0.
References
- 1.Voorhees RM, Ramakrishnan V. Structural basis of the translational elongation cycle. Annu. Rev. Biochem. 2013;82:203–236. doi: 10.1146/annurev-biochem-113009-092313. [DOI] [PubMed] [Google Scholar]
- 2.Wilson DN. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 2014;12:35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- 3.Vázquez-Laslop N, Mankin AS. Context-Specific Action of Ribosomal Antibiotics. Annu. Rev. Microbiol. 2018;72:185–207. doi: 10.1146/annurev-micro-090817-062329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moazed D, Noller HF. Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S ribosomal RNA. Biochimie. 1987;69:879–884. doi: 10.1016/0300-9084(87)90215-x. [DOI] [PubMed] [Google Scholar]
- 5.Gaynor M, Mankin AS. Macrolide antibiotics: binding site, mechanism of action, resistance. Curr. Top. Med. Chem. 2003;3:949–961. doi: 10.2174/1568026033452159. [DOI] [PubMed] [Google Scholar]
- 6.Skinner R, Cundliffe E, Schmidt FJ. Site of action of a ribosomal RNA methylase responsible for resistance to erythromycin and other antibiotics. J. Biol. Chem. 1983;258:12702–12706. [PubMed] [Google Scholar]
- 7.Champney WS, Chittum HS, Tober CL. A 50S ribosomal subunit precursor particle is a substrate for the ErmC methyltransferase in Staphylococcus aureus cells. Curr. Microbiol. 2003;46:453–460. doi: 10.1007/s00284-002-3901-8. [DOI] [PubMed] [Google Scholar]
- 8.Pokkunuri I, Champney WS. Characteristics of a 50S ribosomal subunit precursor particle as a substrate for ermE methyltransferase activity and erythromycin binding in Staphylococcus aureus. RNA Biol. 2007;4:147–153. doi: 10.4161/rna.4.3.5346. [DOI] [PubMed] [Google Scholar]
- 9.Cundliffe E. How antibiotic-producing organisms avoid suicide. Annu. Rev. Microbiol. 1989;43:207–233. doi: 10.1146/annurev.mi.43.100189.001231. [DOI] [PubMed] [Google Scholar]
- 10.McGUIRE JM, et al. Ilotycin, a new antibiotic. Antibiot. Chemother. 1952;2:281–283. [PubMed] [Google Scholar]
- 11.Oliynyk M, et al. Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat. Biotechnol. 2007;25:447–453. doi: 10.1038/nbt1297. [DOI] [PubMed] [Google Scholar]
- 12.Graham MY, Weisblum B. 23S ribosomal ribonucleic acid of macrolide-producing streptomycetes contains methylated adenine. J. Bacteriol. 1979;137:1464–1467. doi: 10.1128/jb.137.3.1464-1467.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonnefoy A, Girard AM, Agouridas C, Chantot JF. Ketolides lack inducibility properties of MLS(B) resistance phenotype. J. Antimicrob. Chemother. 1997;40:85–90. doi: 10.1093/jac/40.1.85. [DOI] [PubMed] [Google Scholar]
- 14.Sutcliffe JA. Antibiotics in development targeting protein synthesis. Ann. N. Y. Acad. Sci. 2011;1241:122–152. doi: 10.1111/j.1749-6632.2011.06323.x. [DOI] [PubMed] [Google Scholar]
- 15.Brockman H, Henkel W. Pikromycin, ein neues Antibiotikumaus Actinomyceten. Naturwissenschaften. 1950;37:138–139. [Google Scholar]
- 16.Almutairi MM, et al. Resistance to ketolide antibiotics by coordinated expression of rRNA methyltransferases in a bacterial producer of natural ketolides. Proc. Natl. Acad. Sci. USA. 2015;112:12956–12961. doi: 10.1073/pnas.1512090112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 18.Theveneau P, et al. The Upgrade Programme for the Structural Biology beamlines at the European Synchrotron Radiation Facility – High throughput sample evaluation and automation. J. Phys. Conf. Ser. 2013;425:012001. [Google Scholar]
- 19.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schluckebier G, Zhong P, Stewart KD, Kavanaugh TJ, Abad-Zapatero C. The 2.2 A structure of the rRNA methyltransferase ErmC’ and its complexes with cofactor and cofactor analogs: implications for the reaction mechanism. J. Mol. Biol. 1999;289:277–291. doi: 10.1006/jmbi.1999.2788. [DOI] [PubMed] [Google Scholar]
- 22.Demirci H, et al. Structural rearrangements in the active site of the Thermus thermophilus 16S rRNA methyltransferase KsgA in a binary complex with 5’-methylthioadenosine. J. Mol. Biol. 2009;388:271–282. doi: 10.1016/j.jmb.2009.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu L, et al. Solution structure of an rRNA methyltransferase (ErmAM) that confers macrolide-lincosamide-streptogramin antibiotic resistance. Nat. Struct. Biol. 1997;4:483–489. doi: 10.1038/nsb0697-483. [DOI] [PubMed] [Google Scholar]
- 24.Pulicherla N, et al. Structural and functional divergence within the Dim1/KsgA family of rRNA methyltransferases. J. Mol. Biol. 2009;391:884–893. doi: 10.1016/j.jmb.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keegan RM, et al. Recent developments in MrBUMP: better search-model preparation, graphical interaction with search models, and solution improvement and assessment. Acta Crystallogr D Struct Biol. 2018;74:167–182. doi: 10.1107/S2059798318003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Terwilliger TC, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The PyMOL Molecular Graphics System, Version 2.3 Schrödinger, LLC.
- 32.Ishida T, Kinoshita K. PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007;35:W460–4. doi: 10.1093/nar/gkm363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 34.Holm L, Laakso LM. Dali server update. Nucleic Acids Res. 2016;44:W351–5. doi: 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rossmann MG, Moras D, Olsen KW. Chemical and biological evolution of nucleotide-binding protein. Nature. 1974;250:194–199. doi: 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- 36.Cheng X, Roberts RJ. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001;29:3784–3795. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bussiere DE, et al. Crystal structure of ErmC’, an rRNA methyltransferase which mediates antibiotic resistance in bacteria. Biochemistry. 1998;37:7103–7112. doi: 10.1021/bi973113c. [DOI] [PubMed] [Google Scholar]
- 38.Maravić G, Bujnicki JM, Feder M, Pongor S, Flögel M. Alanine-scanning mutagenesis of the predicted rRNA-binding domain of ErmC’ redefines the substrate-binding site and suggests a model for protein-RNA interactions. Nucleic Acids Res. 2003;31:4941–4949. doi: 10.1093/nar/gkg666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wass MN, Kelley LA, Sternberg MJE. 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res. 2010;38:W469–73. doi: 10.1093/nar/gkq406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malone T, Blumenthal RM, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 1995;253:618–632. doi: 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- 41.Marchler-Bauer A, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.El-Gebali S, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Farrell HC, Scarsdale JN, Rife JP. Crystal structure of KsgA, a universally conserved rRNA adenine dimethyltransferase in Escherichia coli. J. Mol. Biol. 2004;339:337–353. doi: 10.1016/j.jmb.2004.02.068. [DOI] [PubMed] [Google Scholar]
- 44.Punekar AS, Liljeruhm J, Shepherd TR, Forster AC, Selmer M. Structural and functional insights into the molecular mechanism of rRNA m6A methyltransferase Rlm. J. Nucleic Acids Res. 2013;41:9537–9548. doi: 10.1093/nar/gkt719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X, et al. Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature. 2016;534:575. doi: 10.1038/nature18298. [DOI] [PubMed] [Google Scholar]
- 46.Mendel M, et al. Methylation of Structured RNA by the m6A Writer METTL16 Is Essential for Mouse Embryonic Development. Mol. Cell. 2018;71:986–1000.e11. doi: 10.1016/j.molcel.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doxtader KA, et al. Structural Basis for Regulation of METTL16, an S-Adenosylmethionine Homeostasis Factor. Mol. Cell. 2018;71:1001–1011.e4. doi: 10.1016/j.molcel.2018.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruszkowska A, Ruszkowski M, Dauter Z, Brown JA. Structural insights into the RNA methyltransferase domain of METTL16. Sci. Rep. 2018;8:5311. doi: 10.1038/s41598-018-23608-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boehringer D, O’Farrell HC, Rife JP, Ban N. Structural insights into methyltransferase KsgA function in 30S ribosomal subunit biogenesis. J. Biol. Chem. 2012;287:10453–10459. doi: 10.1074/jbc.M111.318121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tu C, et al. Structural basis for binding of RNA and cofactor by a KsgA methyltransferase. Structure. 2009;17:374–385. doi: 10.1016/j.str.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vester B, Nielsen AK, Hansen LH, Douthwaite S. ErmE methyltransferase recognition elements in RNA substrates. J. Mol. Biol. 1998;282:255–264. doi: 10.1006/jmbi.1998.2024. [DOI] [PubMed] [Google Scholar]
- 52.Bhujbalrao R, Anand R. Deciphering Determinants in Ribosomal Methyltransferases That Confer Antimicrobial Resistance. J. Am. Chem. Soc. 2019;141:1425–1429. doi: 10.1021/jacs.8b10277. [DOI] [PubMed] [Google Scholar]
- 53.Armougom F, et al. Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res. 2006;34:W604–8. doi: 10.1093/nar/gkl092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–4. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–33. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates of ErmE have been deposited in the Protein Data Bank with accession code 6NVM.