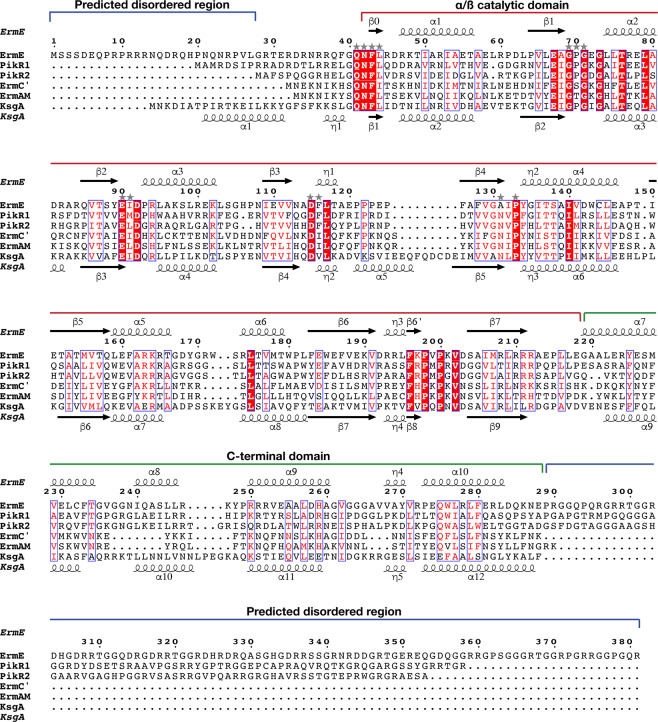
Figure 2.
Structure-guided sequence alignment of ErmE with PikR1, PikR2, ErmC’ (PDB 1QAM)21, ErmAM (PDB 1YUB)23 and KsgA from B. subtilis (PDB 6IFT)52. Domain organization and secondary structure elements of ErmE are shown above the alignment and secondary structure of KsgA below the alignment. Conserved residues are highlighted with white text on red background and conservative substitutions are presented by red text on white background. Residues predicted to interact with SAM are indicated with stars. The alignment was done with Expresso53 and visualized with ESPript 3.054. Disordered regions of ErmE were predicted with PrDOS32.