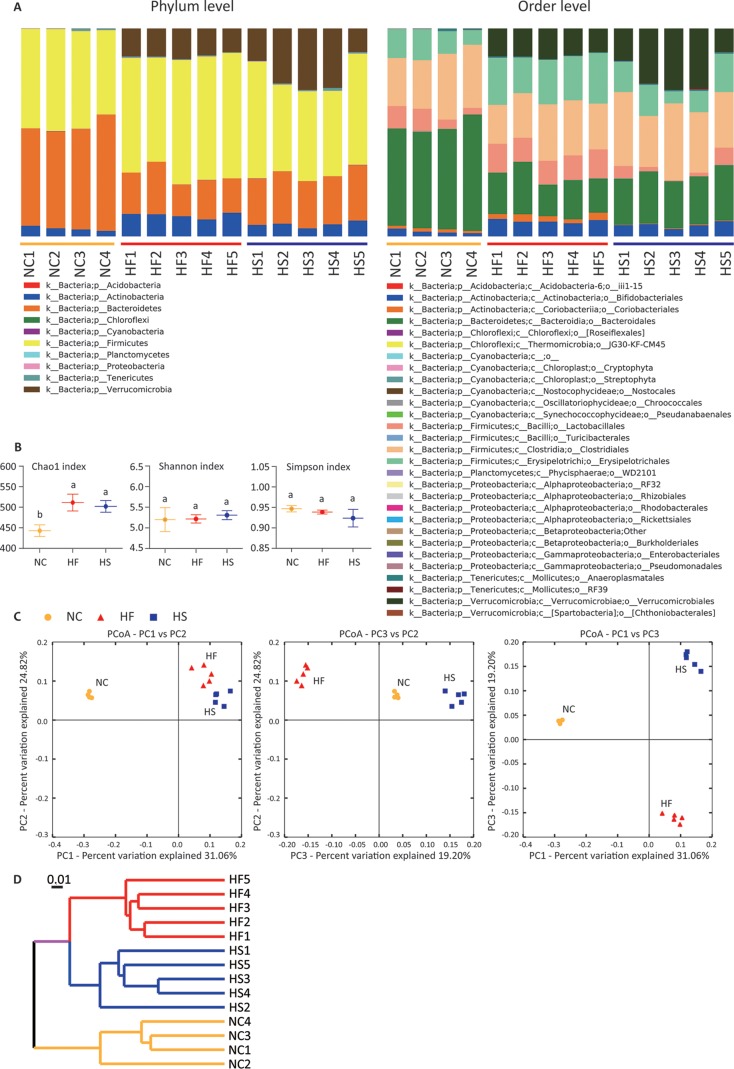
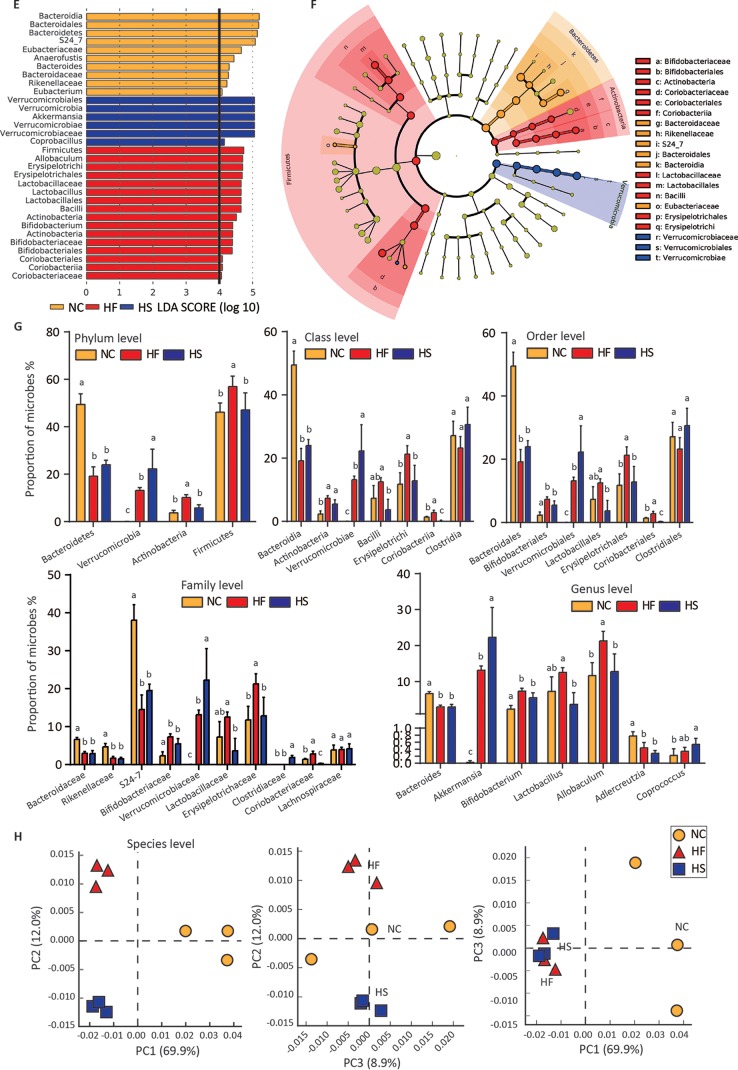
FIG 2.
Different fat-to-sugar ratios shape distinct gut microbiota in prediabetic mice. (A) Phylum- and order-level taxonomic distributions of fecal microbiota from mice fed different diets for 40 days. Stacked columns show the mean of abundance of a given genus as a percentage of the total bacterial sequences in the corresponding group. (B) Analysis of alpha-diversity in mice fed NC, HF, and HS diets diversity by Chao1, Shannon, and Simpson estimators. Groups with different letters have significant differences (one-way ANOVA with Bonferroni test, P < 0.05). (C) Principal-coordinate analysis (PCoA) of samples based on relative abundance of OTUs using weighted UniFrac metric (R2 = 0.84756). (D) Weighted UniFrac tree analysis of samples. The hierarchical clustering structure helps to determine the similarity of the microbial communities between different groups. (E) Linear discriminative analysis (LDA) effect size (LefSe) analysis among the NC (orange), HF (red), and HS (blue) diets. (F) Cardiogram showing differentially abundant taxonomic clades with an LDA score of >4.0 among cases with a P value of <0.05. (G) Relative abundances of bacterial taxa at the different levels. Groups with different letters have significant differences (one-way ANOVA with Tukey test, P < 0.05). (H) Metagenomics revealed microbial composition at the species level. Shown is principal-component analysis of the matrix data for species analysis from mice fed different diets. Accordingly, mice that received the same diet can be obviously clustered. In bar charts, data are shown as means ± SDs.