Abstract
Advances in microarray, RNA‐seq and omics techniques, thousands of long non‐coding RNAs (lncRNAs) with unknown functions have been discovered. LncRNAs have presented a diverse perspective on gene regulation in diverse biological processes, especially in human immune response. Macrophages participate in the whole phase of immune inflammatory response. They are able to shape their phenotype and arouse extensive functional activation after receiving physiological and pathological stimuli. Emerging studies indicated that lncRNAs participated in the gene regulatory network during complex biological processes of macrophage, including macrophage‐induced inflammatory responses. Here, we reviewed the existing knowledges of lncRNAs in the processes of macrophage development and polarization, and their roles in several different inflammatory diseases. Specifically, we focused on how lncRNAs function in macrophage, which might help to discover some potential therapeutic targets and diagnostic biomarkers.
Keywords: cancer, immune response, inflammation, long non‐coding RNA, macrophage polarization
1. INTRODUCTION
Macrophage plays a crucial intermediary role in human immune response. They can be activated after innate immunity receptors sensing the tissue damage and metabolic dysfunction, which is considered as the first line of defence for human immunity.1, 2 In response to various stimuli, macrophages have a strong functional and phenotypic plasticity,3 and can be polarized to the classical M1 type and the M2 type.4 Toll‐like receptor (TLR) ligands, macrophage receptors with collagenous structure (MARCO) and interferon γ (IFN‐γ) may induce M1 macrophages and then promote the production of some pro‐inflammatory mediators including tumour necrosis factor‐α (TNF‐α), cytokines IL‐1, IL‐6 and IL‐23, inducible nitric oxide synthase (iNOS) and HLA‐DR.5 However, IL4/IL13 (M2a), immune complex (M2b), and the anti‐inflammatory cytokine IL‐10 or transforming growth factor‐β (M2c) can induce M2 macrophages and produce IL‐10, mammalian chitinase Ym1, arginase 1 (Arg1), resistin‐like molecule α1 (Fizz1), CD163 and chitotriosidase against inflammation.6, 7, 8 The signalling pathways involved in macrophage activation follow the common four pathways, including JNK, JAK/STAT, Notch and PI3K/Akt pathways.8, 9 Furthermore, macrophages can experience phenotypic conversion to adapt to some specific environmental changes,10 such as cancer,11 fatty liver,12 tissue repair and remodelling.13
Next‐generation RNA sequencing and omics techniques have displayed that most of the human genomes were transcribed into RNAs, and nearly 98% of them without coding for proteins. These 98% of RNAs are called non‐coding RNAs (ncRNAs).14 LncRNAs are ncRNAs with critical biological roles and a length of more than 200 nucleotides.15, 16, 17 The mechanisms of lncRNAs remain unclear,18 but increasing number of in vitro and in vivo studies have shown that lncRNAs can be expressed in macrophage during its development and control some gene expression.19, 20, 21 However, a systematic review uncovered the interactions among lncRNAs expressed in macrophage is not available. Hence, we reviewed recent advances in elucidation of the variability of lncRNA function within macrophages, and mainly focused on the following four aspects: biology of lncRNAs, involvement of lncRNAs in human monocyte/macrophage differentiation, characteristics of lncRNAs in macrophage polarization and dysregulation of lncRNAs in macrophage involved inflammatory diseases. Overall, this review may help to discover some potential therapeutic targets and diagnostic biomarkers for multiple diseases.22, 23
2. BIOGENESIS, STRUCTURES AND FUNCTION MANNERS OF LNCRNAS
Compared with other ncRNAs, the mechanisms of lncRNAs were still unclear because of its complexity and relatively poor conservation.24 Increased researches have already extensively enhanced the knowledge of lncRNAs, and some articles have processed stemmatic reviews on its biology, structures and function manners. For details, see Beermann,14 Fatica25 or Quinn.26 Briefly, biogenesis of lncRNAs are usually transcribed by RNA Pol II(RNPII) in the nucleus, and only a fraction are more likely to be transcribed by RNPIII.24 LncRNAs can be transcribed from intergenic (between protein‐coding genes, lincRNA), intronic, natural antisense transcripts (NATs) or promoters and enhancers.27 Bio‐functional activities of lncRNAs rely on base pairing upon the primary structure, or develop by higher‐order configurations on the basis of the secondary structures.28, 29 Diversity of their structures enabled lncRNAs to perform a variety of functions.30 LncRNAs share the same cap and polyadenylate tail structures with mRNAs, but express lower levels than mRNAs, and in generally exhibit more specific expression profiles and precise expression patterns.31, 32
The regulation of lncRNAs is also complex and diverse. Most transcribed lncRNAs tend to remain in the cell nucleus and be assembled into chromosomes during biological processes.31 Biological functions of most lncRNAs in accordance with previous reports can be primarily classified into 3 types14, 33: (a) LncRNAs act as regulator of genomic transcription in the nucleus. Transcription of certain genes are regulated by binding chromatin‐modifying factors, heterogeneous nuclear ribonucleoprotein (hnRNPs) or transcription factors, with cis‐ or trans‐acting lncRNAs and enhancer RNAs (eRNAs). Cis‐regulatory way may directly silence the nearby genes of transcribed place by methylation of histone H3 and so on.34 However, most lncRNAs work in a trans‐regulatory way and play a repressor or activator role in some distant gene loci which are not even on the same chromosome.35 LncRNAs can also originate from enhancer elements called eRNAs, acting on the regulation of enhancer activity.36 (b) LncRNAs participate in post‐transcriptional moderation in the cytoplasm. These lncRNAs can promote or weaken the translation of target mRNAs, even alter the stability of mRNAs and proteins, or change the protein translocation.29, 33, 37, 38 Moreover, they can function as competing endogenous RNAs (ceRNAs), also called microRNA (miRNA) sponges, to protect the target mRNAs expression by directly binding to miRNAs or keeping miRNAs away from mRNAs.39 (c) The last function manner of lncRNAs is secreted to extracellular. It has been displayed that lncRNAs could be packed in extracellular vesicles (EVs), such as exosomes, secreted out of cell alone or bound to proteins.40 Circulating lncRNAs show advantages in biomarkers, regulating many pathophysiological processes41 (shown in Figure 1).
Figure 1.

Biogenesis, structures and functions of lncRNAs. EBF: enhancer‐binding factors. The biogenesis of lncRNAs is mainly transcribed by RNPII in the nucleus. Bio‐functional activities of lncRNAs rely on base pairing upon the primary structure, or develop by higher‐order configurations on the basis of the secondary structures including helices, hairpin loops, bulges and pseudoknots. Biological functions of lncRNAs can be primarily classified into 3 types: acting as regulator of genomic transcription in the nucleus via cis‐ or trans‐acting lncRNAs and eRNAs; participating in post‐transcriptional moderation in the cytoplasm and involving in peripheral circulation.28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41 LncRNAs promoted biological processes showed in green arrow, and negative regulated displayed in red line
3. LNCRNAS INVOLVE IN HUMAN MONOCYTE/MACROPHAGE DIFFERENTIATION
Monocytes/macrophages originated from haematopoietic stem cells (HSCs) play a major role in non‐specific immunity and inflammatory response.42 Monocyte is a population of mononuclear leucocyte in circulating blood cells. It can transfer to diverse tissues and then differentiate into macrophage. This complex differentiation process requires a coordinated expression of transcription factors, cytokines and ncRNAs.43 Although many of well‐defined miRNAs have been recognized as key regulators involved in haematopoietic differentiation, there were few reports about lncRNAs regulating monocyte/macrophage differentiation.44 Chen et al have revealed that phorbol‐12‐myristate‐13‐acetate (PMA) could raise the long non‐coding monocytic RNA (lnc‐MC) expression in human THP‐1 cell line, HL‐60 cell line and CD34+ haematopoietic stem‐progenitor cells (HSPCs). Mechanically, lnc‐MC acted as a ceRNA to soak up miR‐199a‐5p and release activin A receptor type 1B (ACVR1B),45 then enhanced the effect of haematopoiesis‐specific transcription factor PU.1 on suppressing miR‐199a‐5p and eventually facilitated the differentiation process through activating the transforming growth factor β (TGF‐β).46 Once monocyte recruiting from bone marrow to peripheral blood, it can differentiate into macrophage with phenotypic variability.47 LncRNA NTT was found to be expressed in resting human primary monocyte, monocyte‐derived macrophage and the THP‐1 cell line. Yang et al have elucidated the role of lncRNA NTT in monocyte after knocking down it in THP‐1 cells. Briefly, the key transcription factor of monocyte C/EBPβ bound to the NTT promoter and regulated the NTT expression, which might enhance the PBOV1 expression by interacting with hnRNPU and the promoter of PBOV1. In lipopolysaccharide (LPS)‐treated THP‐1 cells and peripheral blood mononuclear cells (PBMCs) of first‐diagnosed untreated early rheumatoid arthritis (RA) patients, the C/EBPβ/NTT/PBOV1 axis was found to be hyperactivated. Increased expression of PBOV1 resulted in cell cycle G1 arrest, differentiation into macrophages, high IL‐10 and CXCL10 mRNA levels, and up‐regulation of the costimulatory molecules48 (Figure 2 and Table 1).
Figure 2.
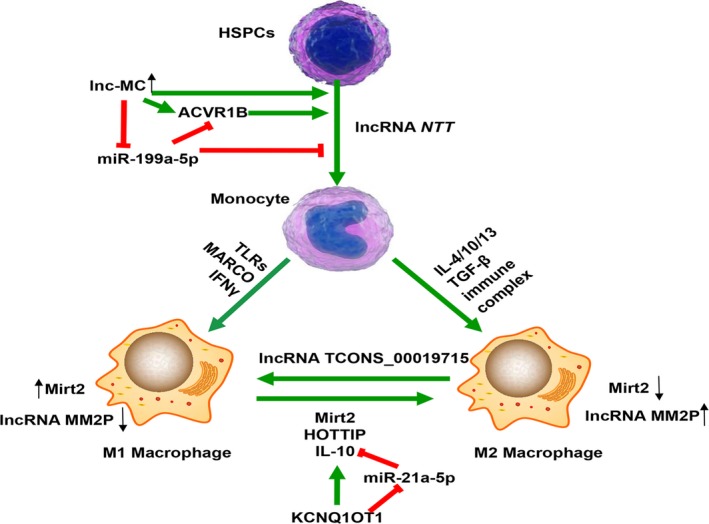
Involvement of lncRNAs in monocyte/macrophage development and M1/M2 switch. Circulating monocytes originated from haematopoietic stem/progenitor cells (HSPCs) are considered as the precursors of macrophages. Induced by phorbol‐12‐myristate‐13‐acetate (PMA), lnc‐MC up‐regulated in THP‐1, HL‐60 cells and CD34+ HSPCs. It could act as a ceRNA to soak up miR‐199a‐5p, and eventually facilitate the differentiation process through activating the transforming growth factor β (TGF‐β) signalling pathway. 45, 46 In response to various stimuli, macrophages mainly polarized into two types: classical M1 and alternative M2 type, and dynamically switched its function to adapt to the specific environmental changes. LPS induced up‐regulation of lncRNA Mirt2 in the cytoplasm, but the increase in Mirt2 was restricted by sustained and excessive activation of inflammatory responses at the late stage. Restoring Mirt2 expression in later stage promoted the IL‐4 induced M2 polarization with a remarkable increased level of M2 markers.53 HOTTIP expression was associated with TLR tolerance, and has been speculated to skew macrophage polarization to a “M2‐like” phenotype.82 KCNQ1OT1 could function as a miR‐21a‐5p decoy to up‐regulate IL‐10, induced polarization of macrophages into M2 type.83, 84 LncRNA TCONS_00019715 was expressed at a higher level when M2 macrophage was converted to M1, but decreased when M1 type was converted to M285
Table 1.
Main mechanisms of lncRNAs expressed in macrophages during development and polarization
| Phases | Positive regulated | Mechanisms and models | Refs | Negative regulated | Mechanisms and models | Refs |
|---|---|---|---|---|---|---|
| Monocyte/macrophage differentiation | lnc‐MC | Act as miR‐199a‐5p sponge, releases it suppressor, THP‐1, HL‐60 cells and CD34+ HSPCs | 45, 46 | ‐ | ||
| M1‐like macrophages induced by inflammatory response | lncRNA‐Nfkb2, lncRNA‐Rel | Unknown, BMDMs | 57 | lncRNA Lethe | Keep p65 subunit away from DNA and inhibit inflammatory genes expression. NOX2 expression and ROS production, RAW264.7cells | 71 |
| AS‐IL1α | Recruit RNPII to the IL‐1α promoter, and elevate H3K9, BMDMs | 58 | lincRNA‐p21 | Sequester p65 mRNA and attenuate the translation of p65, human Jurkat T cell and THP‐1 monocyte lines | 72 | |
| lncRNA cox‐2(PACER) | Interacte with the p50/p50 homodimeric and act in cis‐ as a decoy to activate the promoter of cox‐2, human and mouse macrophages | 49, 59 | lnc‐IL7R | Enhance trimethylation of H3K27, lead to transcriptional silence, THP‐1 | 73 | |
| IL1β‐eRNA, IL1β‐RBT46 | Act as eRNAs to promote the expression of IL1‐β and CXCL8, THP‐1 | 60 | lincRNA THRIL | Interact with hnRNPL at the promoter of Tnfα to make sure the expression in dose control, human macrophages | 74, 75 | |
| lincRNA‐Tnfaip3 | Interact with the HMGB1 protein and facilitate Hmgb1‐associated histone modification, murine macrophages | 61, 62 | lncRNA SeT | Attenuate the stability of Tnfα mRNA levels, murine macrophages | 20 | |
| lncRNA‐CCL2 | Suppress histone deacetylase Sirtuin‐1, sepsis mice and LPS‐ stimulated macro phages | 64 | NKILA | Modify the phosphorylation motifs of IĸBα to block its degradation, breast cancer cell lines | 77 | |
| lncRNA FIRRE | Interact with hnRNPU and enhance stability of several inflammatory mRNAs, human macrophages and intestinal epithelial cells | 65 | lincRNA‐EPS | Function as a scaffold to bind hnRNPL, BMDMs | 78 | |
| lncRNA SNHG16 | Act as a ceRNA to up‐regulate TLR4 by down‐regulating the miR‐15a/16 cluster, RAW264.7 cells | 66 | MALAT1 | Interact with NF‐κB in the nucleus, keep it away from DNA and decrease inflammatory cytokines, THP‐1, RAW264.7 cells | 79 | |
| lincRNA‐Cox2 | Assemble into SWI/SNF complex; affecting IκBα degradation in the cytoplasm; interact with hnRNP‐A2/B1 and hnRNP‐A/B to suppress inflammation; promote recruitment of Mi‐2/NuRD repressor complex, which led to increased H3K27 dimethylation at Il12b promoter, BMDMs | 67, 68, 69, 70 | MacORIS | A repressor of IFN‐γ signalling, human THP‐1 macrophages | 81 | |
| lncRNA MALAT1 | Suppression of inflammatory responses by up‐regulating miR‐146a, murine alveolar macrophage cell line MH‐S | 80 | ||||
| M2‐like macrophages | lncRNA Mirt2 | Inhibit TRAF6 oligomerization and ubiquitination, RAW264.7 cells | 53 | ‐ | ||
| HOTTIP | Unknown, endotoxin‐tolerized macrophages | 50 | ‐ | |||
| lncRNA KCNQ1OT1 | Function as a miR‐21a‐5p decoy to up‐regulate IL‐10, PMMA‐induced BMDMs. | 83, 84 | ‐ | |||
| M1/M2 switch | lncRNA TCONS_00019715 | Decrease when M1 type is converted to M2, THP‐1, MDMs | 85 | PAK1 | Be helpful to M1 macrophage polarization, THP‐1, MDMs | 85 |
Abbreviations: BMDMs, bone marrow‐derived macrophages; H3K9, acetylation of histone H3 at lysine 9; HMGB1, high‐mobility group box 1 protein; hnRNPU, heterogeneous nuclear ribonucleoprotein; MDMs, monocyte‐derived macrophages; Mi‐2/NuRD, Mi‐2/nucleosome remodelling and deacetylase; NOX2, NADPH oxidase 2; PMMA, polymethyl methacrylate; ROS, reactive oxygen species; SWI/SNF, SWItch/Sucrose Non‐Fermentable; TRAF6, TNF receptor‐associated factor 6.
4. LNCRNAs EXPRESSION IN MACROPHAGE POLARIZATION AND FUNCTION
4.1. lncRNAs expression profile in M1‐like macrophage
Granulocyte macrophage colony stimulating factor (GM‐CSF), LPS and Toll‐like receptor (TLR) ligands always contribute to M1 polarization in macrophage. That macrophage might be polarized through the JNK pathway after binding to CCR2 and nuclear factor‐κB (NF‐κB), or through the activation of PI3K with increased RelA/NF‐κB activity.8 Functionally, M1‐type macrophage promotes Th1 response and produces copious amounts of pro‐inflammatory cytokines or reactive oxygen species (ROS) to kill pathogens.44 TLR‐triggered NF‐κB is one of the best studied pathways participated in the conversion of macrophage to M1‐like phenotype,49 which is shown in Figure 3.50, 51, 52, 53 All TLRs, excluding TLR3, mainly activate the NF‐κB‐dependent or IRF7‐dependent type I IFN (TLR7‐9) pro‐inflammatory signallings via the adapter protein myeloid differentiation marker 88 (MyD88).54, 55 Many transcriptome sequencing analyses of macrophages stimulated with TLR ligands have shown that numerous genes and lncRNAs involved in macrophage polarization (Figure 3 and Table 1), and these macrophages tended to be induced into M1 type, along with high levels of pro‐inflammatory mediators.56
Figure 3.
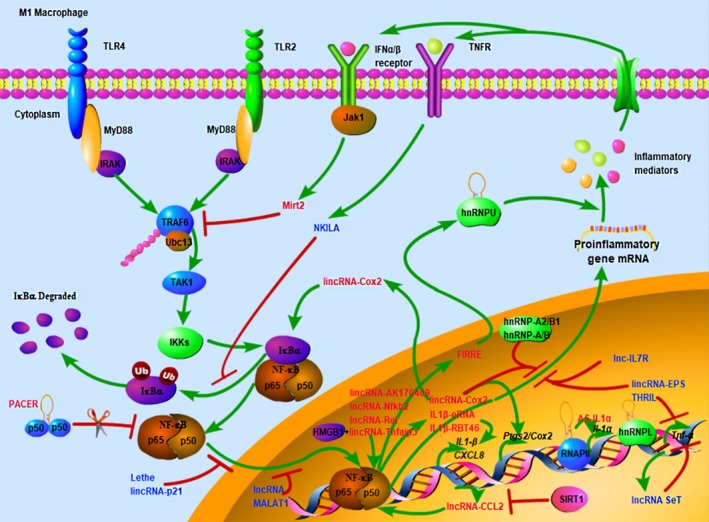
Involvement and regulation of lncRNAs in M1‐like macrophage polarization. Many toll‐like receptor (TLR)‐triggered polarization of M1‐like macrophage mainly depends on nuclear factor‐κB (NF‐κB) signalling pathway. At the plasma membrane, the binding of myeloid differentiation marker 88 (MyD88) to TLRs results in the recruitment and phosphorylation of IL‐1 receptor‐associated kinases (IRAKs), which facilitates oligomerization (via K63) and auto‐ubiquitination (via E2 ubiquitin ligase Ubc13) of TNF receptor‐associated factor 6 (TRAF6). Ubiquitinated TRAF6 subsequently activates other signalling proteins, such as transforming growth factor β‐activated kinase (TAK1). Then TAK1 activates the inhibitor of κB α (IκBα) kinases (IKKs), leading to degradation of IκBα and activation of NF‐κB, immune and inflammatory responses. LncRNAs promote the development of M1 macrophage via NF‐κB or other pathways showed in red font, whereas negative regulated displayed in blue font50, 51, 52, 53
In response to TLR4 (LPS) activation, lncRNA‐Nfkb2 and lncRNA‐Rel, located near some classical pro‐inflammatory transcription factors, were up‐regulated and co‐regulated with inflammatory response in mouse bone marrow‐derived macrophages (BMDMs).57 AS‐IL1α was a NAT located in the nucleus of macrophage and expressed at a low level in resting cells. It was highly up‐regulated after stimulating by TLRs via NF‐ĸB signal, and then recruited RNPII to the IL‐1α promoter. In the presence of abundant AS‐IL1α, elevated acetylation of histone H3 at lysine 9 (H3K9) promoted the combination of RNPII and the promoter of IL‐1α, but it could not control the gene's expression. 58 LncRNA cox‐2 (also has been called PACER) was located at upstream of the cyclooxygenase‐2 (cox‐2) gene in both human and murine macrophages. PACER could directly interact with the p50/p50 homodimeric, an inhibitor of NF‐κB family and subsequently resulted in activation of the cox‐2 promoter.49, 59 Moreover, lnc IL1β‐eRNA and IL1β‐RBT46 were found to be transcribed from enhancer regions via NF‐ĸB in THP‐1 cells, and predominantly localized to the nucleus, functioning as eRNAs to promote the expression of IL1‐β and CXCL8.60 After LPS stimulation, lincRNA‐Tnfaip3, which is located at the tumour necrosis factor a‐induced protein 3 (Tnfaip3) gene, mediated early response through NF‐κB signalling in murine macrophages. LincRNA‐Tnfaip3 could non‐covalently unite to the high‐mobility group box 1 protein (HMGB1), DNA and RNA molecules, then assembled into a complex to facilitate Hmgb1‐associated histone modification and the combination of DNA and p65/p50, eventually promoted the transcription of many inflammatory and defence genes in macrophages,61 as well as other mice tissues induced by LPS.62 Sirtuin‐1 (SIRT1) is a conserved histone deacetylase, which can inhibit macrophage response by deacetylating H3K9 in lncRNA‐CCL2 loci and transcriptionally suppressing the lncRNA‐CCL2 level.63 LncRNA‐CCL2 was significantly up‐regulated in sepsis mice or LPS‐stimulated macrophages, but the SIRT1 expression was down‐regulated.64 Lu et al have reported that increased binding of p65 to the promoter of functional intergenic repeating RNA element (FIRRE) gene mediated lncRNA FIRRE up‐regulation in macrophages and intestinal epithelial cells after LPS stimulation. FIRRE was a NF‐κB‐controlled lncRNA and specifically interacted with hnRNPU, then regulated the mRNA stability of several genes by binding to the AU‐rich elements of these mRNA.65 LncRNA SNHG16 was released into cell cytoplasm and could act as a ceRNA of miR‐15a/16 to up‐regulate TLR4 in RAW264.7 cells.66
TLR2 activation could induce a high lincRNA‐Cox2 level and promote its neighbouring prostaglandin‐endoperoxide synthase 2 (Ptgs2/Cox2) gene expression in BMDMs through MyD88/NF‐κB signalling pathway, mediating an early inflammatory gene production.67 After lincRNA‐Cox2 being recruited to the SWItch/Sucrose Non‐Fermentable (SWI/SNF) complex in macrophages, NF‐κB subunits subsequently bound to this complex, finally triggered SWI/SNF‐associated chromatin remodelling and transcription of late inflammatory genes.68 A RNA‐seq analysis in TLR‐activated BMDMs indicated that lincRNA‐Cox2 and lincRNA‐AK170409 were involved in controlling NF‐κB signalling. Unlike lincRNA‐Cox2 causing IκBα degradation in cell cytoplasm, lincRNA‐AK170409 was only retained in the nucleus and up‐regulated various inflammation genes by affecting other components of NF‐κB.69 Meanwhile, lincRNA‐Cox2 could act as a suppressor of inflammation by directly interacting with the hnRNP‐A2/B1 and hnRNP‐A/B.67 Additionally, in intestinal epithelial inflammatory, lincRNA‐Cox2 significantly decreased the transcription of Il12b (a late‐responsive cytokine) by mobilizing the Mi‐2/nucleosome remodelling and deacetylase (Mi‐2/NuRD) repressor complex to the promoter region of Il12b, which led to increased H3K27 dimethylation at the promoter and suppressive expression of Il12b.70
LncRNA Lethe, lincRNA‐p21 and lnc‐IL7R were increased in macrophages activated through NF‐κB pathway, and acted in a negative feedback loop to inhibit NF‐κB‐induced inflammation. Mechanically, lncRNA Lethe combined with the p65 subunit of NF‐κB and kept it away from DNA, and then presented an anti‐inflammatory effect by blocking the activation of downstream inflammatory genes, NADPH oxidase 2 (NOX2) expression and ROS production.71 LincRNA‐p21 was also found to attenuate the p65 mRNA and its translation, leading to TNF‐α‐stimulated NF‐ĸB activity.72 Increased lnc‐IL7R was initially expressed in the nucleus and likely inhibited MyD88‐dependent LPS‐inducible genes expression by enhancing the trimethylation of H3K27, a mark of transcriptional silence, at its promoters. However, it did not seem to involve in regulating the expression of overlapped IL7R gene and viral infection.73 Similarly, lincRNA THRIL as a negative regulation of NF‐κB could interact with hnRNPL at the Tnfα gene and ensure the expression of TNF‐α in dosage control, but it down‐regulated in human macrophages after TLR2 activation.74, 75 Encoded by the Tnfα loci, lncRNA SeT increased in LPS‐treated murine macrophage,76 with unstable Tnfα mRNA.20 The above lncRNAs are important to reduce TNFα expression in macrophages. NKILA was highly expressed to restrain the NF‐ĸB‐driven inflammation in response to TNF‐α irritant. NKILA could decorate the phosphorylation of IĸBα to inhibit its degradation and prevent NF‐ĸB from being transferred to the nucleus.77 LincRNA‐EPS in resting macrophages, which inversely controls the level of IL‐6, was overexpressed but reduced via TLR ligation triggered NF‐ĸB pathway. Atianand et al78 revealed that the recruitment of RNPII and trimethylation of H3K4 near the transcription start site (TSS) of immune response genes were transcriptionally suppressed by lincRNA‐EPS, which functioned as a scaffold to hnRNPL. LncRNA MALAT1 was up‐regulated in LPS‐activated macrophage. In the nucleus, it can combine with NF‐κB and keep its complex from binding DNA, then inhibit the expression of inflammatory cytokines.79 Knocking down the MALAT1 in LPS‐induced murine alveolar macrophage MH‐S showed a lower inflammatory response by up‐regulating the miR‐146a expression.80 LincRNA MacORIS was located in cell cytoplasm, and served as a depressor of IFN‐γ in THP‐1 macrophage, leading to decrease in IFN‐γ–induced Janus kinase 2 (JAK2) and phosphorylation of signal transducer and activator of transcription 1 (STAT1), but that did not happen in mouse macrophages.81
4.2. Characteristics of lncRNAs in M2‐like macrophage, M1/M2 switch or TMAs
Macrophage activated through an opposite manner is known as M2 type. M2 macrophage plays an essential role in adaptive response and contributes to Th2‐type response involved in acute tissue damage and repair.44, 47 However, studies on the expression of lncRNAs in M2 macrophage are still rare, and the exact mechanism of action is still unclear. Du et al found that lncRNA Mirt2 was up‐regulated in the cytoplasm of macrophage after LPS stimulating, then inhibited the oligomerization and ubiquitination of TNF receptor‐associated factor 6 (TRAF6) and relieved inflammatory responses at the late stage. They speculated that Mirt2 might hide the ubiquitination sites for the E2 ubiquitin ligase Ubc13, and induce a decreased ubiquitination level of TRAF6 (Figure 2 and Table 1). At the early stage, IL‐4 could reduce Mirt2 via the Jak‐Stat6 pathway. After restoring the expression of Mirt2, IL‐4 treated macrophage was inclined to M2 polarization with a high level of M2 molecule markers. However, the elaborate mechanisms of macrophage polarization induced by Mirt2 are still unknown.53 Murphy et al conducted real‐time PCR in LPS‐tolerized macrophage and found increased expression of lncRNA HAR1A and HAR1B but decreased levels of PCGEM1 and HOTTIP lncRNAs.50 Regulating the expression of HOTTIP is associated with TLR tolerance, and has been speculated to promote macrophage into M2 phenotype.82 In polymethyl methacrylate (PMMA)‐induced BMDMs, the expression of TNF‐α, iNOS and miR‐21a‐5p were increased, but the expression of IL‐10, Arg1 and lncRNA KCNQ1OT1 were suppressed. KCNQ1OT1 might act as a miR‐21a‐5p decoy to up‐regulate IL‐10 expression,83 and then prompt macrophages into M2 type.84 A detailed transcriptome analysis in human macrophage revealed that lncRNA TCONS_00019715 was highly expressed when M2 type was converted to M1, but decreased when M1 type was converted to M2. Like TCONS_00019715, overexpressed P21‐activated kinase 1 (PAK1) in macrophage was helpful to M1 macrophage polarization.85 During M2 polarization, lncRNA‐MM2P was increased, which was contrary to M1 macrophage. Knockdown of lncRNA‐MM2P in RAW264.7 could cut down the M2 polarization by decreasing phosphorylation level of STAT6.86 LncRNA growth‐arrest‐specific transcript 5 (GAS5) was up‐regulated in human monocyte‐derived macrophages from pneumonia children and acted as a miR‐455‐5p decoy to up‐regulate SOCS3, then inhibited JAK2/STAT3 signalling and increased the transformation of M1 macrophage87 (Figure 2 and Table 1).
Tumour‐associated macrophages (TAMs) have a particular polarization state which is deemed as M2‐like macrophage.47 Incubated in thyroid tumour microenvironment, macrophage showed an increased expression level of MALAT1, which mediated secretion of fibroblast growth factor‐2 (FGF2) protein, leading to decreased cytokine release, enhanced tumour neovascularization, proliferation of thyroid cancer cells and metastatic potential.3, 88 In TAMs isolated from thyroid cancer, expression of lncRNA nuclear enriched abundant transcript 1 (NEAT1), which is localized in the nucleus (24), and Arg‐1 were obviously elevated, while miR‐214 was significantly decreased. Indirectly, NEAT1 promoted thyroid cancer growth by inducing β‐catenin expression.89 TAM‐derived exosomal miR‐146b‐5p might target with TRAF6 gene, and inhibit endothelial cells migration through decrease in matrix metalloproteinase‐2 (MMP2). After co‐culturing with epithelial ovarian cancer (EOC)‐derived exosomes, this inhibition was reversed remarkably. Additionally, lncRNA ENST00000444164 and ENST0000043768 were overexpressed in EOC‐derived exosome and might regain the migration of endothelial cell via NF‐κB phosphorylation.90 In hepatocellular carcinoma (HCC), lncRNA cox‐2 could suppress the tumorigenesis and metastasis by promoting M1‐type macrophage and inhibiting M2 type.56 LncRNA uc.306 was up‐regulated in U937 cells during M2 differentiating to M1 phenotype. Uc.306 was also low expressed in HCC tissues, indicated a potential prognostic biomarkers of HCC.91 Breast cancer cells showed a high expression of lncRNA urothelial cancer‐associated 1 (UCA1) when cultured with macrophages. UCA1 could promote the proliferation, migration and angiopoiesis of tumour cells, and had a good correlation with the progress of breast cancer.92 Chen et al proved that aerobic glycolytic tumour cells released lactate to up‐regulate the EV‐transmitted lncRNA HISLA in macrophages, which induced chemoresistance and shorter survival of patients with breast cancer by blocking the interaction of PHD2 and HIF‐1α and inhibiting the hydroxylation and degradation of HIF‐1α.93 When co‐cultured with prostate cancer cell line PC‐3 cells, miR‐148a was highly expressed in TAMs, whereas CCAT1 and PKCζ were highest in M1 macrophage. Down‐regulated lncRNA CCAT1 promoted M2 macrophage polarization by up‐regulating miR‐148a, and then down‐regulating the expression of PKCζ.94
In addition to M1, M2 type and TAMs, some macrophage subsets with unique characteristics and functions have also been reported, such as CD169+ or TCR+ macrophage.47 However, it is still not clear about these newly discovered macrophage types, and more research is needed to prove its origin, biomarkers and differentiation processes. Hence, we only reviewed classical M1, M2 macrophage and TAMs.
5. INFLAMMATORY DISEASES INDUCED BY DYSREGULATION OF LNCRNAS AND MACROPHAGES
5.1. Atherosclerosis and cholesterol transport
Macrophages take up oxidized low‐density lipoproteins (oxLDL), causing apoptosis to form foam cells, which is the basis of atherosclerosis.95 A transmembrane protein, the type B scavenger receptor CD36, is up‐regulated on macrophage's cell membrane by oxLDL uptaking during this progress.96 After oxLDL treatment, lncRNA MALAT1 transcription was activated through NF‐κB signal. Numerous expressed lncRNA MALAT1 promoted CD36 expression through recruiting β‐catenin to the promoter region of CD36, and participated in CD36‐mediated lipid uptaking.97, 98 LncRNA MALAT1 expressed in macrophage of diabetic atherosclerosis (DA) was also obviously increased, which might promote the pyroptosis of normal macrophage. Low dose of chronic sinapic acid (SA) could down‐regulate lncRNA MALAT1 in DA rats and release the pyroptosis.99 When THP‐1 cells treated with oxLDL, the expression of lncRNA HOTAIR was significantly elevated, followed by decrease in miR‐330‐5p, which mediate low oxidative stress and inflammation.100 After binding to the E3 ubiquitin‐protein ligase and mouse double minute 2 (MDM2), lincRNA‐p21 could heighten the transcription of p53 by forcing MDM2 released from the p53 (cell cycle and apoptosis control molecule), subsequently allowed p53 binding to p300 and acted on their target genes. But in atherosclerotic plaques, expression of lincRNA‐p21 was depressed.101 LncRNA taurine up‐regulated gene 1 (TUG1) was elevated in HFD‐treated ApoE‐/‐ mice, oxLDL‐induced mouse macrophages and mouse VSMCs. As a target of TUG1, miR‐133a was correspondingly reduced and reversed the expression of fibroblast growth factor 1 (FGF1) protein, further inhibited cell apoptosis and aggravated the foam cell formation.102 Another increased lncRNA H19 in oxLDL‐treated macrophage might facilitate effects on foam cell formation and inflammation response by reducing miR‐130b.103 Hu et al revealed that in THP‐1‐derived foam cells, lncRNA RP5‐833A20.1 (NFIA‐A1) was increased but the transcription factor nuclear factor 1A (NFIA) was decreased. Besides, induced high miR‐382‐5p level might diminish the NFIA mRNA and protein.104 Further reports elucidated that NFIA could promote retrograde cholesterol transport (RCT), reduce the circulating levels of pro‐inflammatory cytokines and significantly improve plasma hyperlipidaemia.105 Transmembrane protein ATP‐binding cassette transporter A1 (ABCA1) takes part in the process of RCT. Human high‐density lipoprotein (HDL) deficiencies, such as syndrome sterol deposits and premature atherosclerosis, are caused by ABCA1 mutations.106 In THP‐1 derived foam cells, lincRNA‐DYNLRB2‐2 was distinctly increased, which promoted ABCA1‐mediated RCT and inhibited inflammation response by up‐regulating G‐protein‐coupled receptor (GPR119), and eventually slowed the atherosclerotic plaque formation down.107 Sallam et al have identified the lncRNA MeXis positive regulated the critical cholesterol efflux gene Abca1, which encodes ABCA1 protein. Lack of MeXis in mouse BMDMs affected the structure of the Abca1 loci, had a negative impact on cholesterol overload and promoted plaque formation. Mechanically, the transcriptional coactivator DDX17 was recruited to the promoter of MeXis and magnified the LXR‐dependent transcription of Abca1.108 Work by Chen et al109 showed that lncRNA GAS5 was highly expressed not only in the plaque of atherosclerosis patients but also in animal models. Some reports indicated that the elevated lncRNA GAS5 might motivate the apoptosis of THP‐1‐derived foam cells,110 whereas other study confirmed that the enriched GAS5 could exacerbate the secretion of pro‐inflammatory cytokines and chemokine induced by oxLDL. Additionally, GAS5 suppressed the miR‐221 expression as a sponge, which might cause plaque destabilization.111 After oxLDL treatment, lncRNA GAS5 was abundant in THP‐1‐derived exosomes and enhanced vascular endothelial cell apoptosis by sucking up the exosomes.112
5.2. Diabetes mellitus
Transcriptome profiling of BMDMs from diabetic db/db mice indicated a pro‐inflammatory, pro‐fibrogenic and disordered polarization of macrophage. A RNA sequencing and real‐time qPCR demonstrated that lncRNA Dnm3os (dynamin 3 opposite strand) was up‐regulated via NF‐κB activation in BMDMs from type 2 diabetic db/db mice as well as monocytes from type 2 diabetic patients. Stable overexpression of Dnm3os in macrophages altered global histone modifications and promoted inflammation, immune response and phagocytosis.113 LncRNAs E330013P06 (E33) is one of the visibly increased lncRNAs in macrophages under T2DM conditions. Compared with db/+ mice, pro‐inflammatory genes were overexpressed from db/db mice, which indicated that diabetes inhibited M2 phenotypic transformation and promoted the inflammatory response of M1 macrophage. Additionally, expression of E33 in macrophages greatly increased the CD36 level, and further supported the foam cell formation and proatherogenic responses of macrophages.114 A new study indicated that, under high glucose conditions or diabetic wounds, lncRNA Lethe was decreased in macrophages, and released more free p65‐NF‐κB to the nucleus, which promoted ROS and NOX2 production, eventually impaired wound healing.115
5.3. Coeliac disease
Coeliac disease (CeD) is a immune‐mediated primary intestinal malabsorption syndrome caused by gluten intolerance, and the pathological feature is small intestinal mucosal lesions.116 LncRNA lnc13 contains a CeD‐associated haplotype block and expresses at a low level in resting cells. A low level of lnc13 was detected in CeD patients’ small intestinal biopsy samples and BMDMs treated with LPS stimulus. The overall expression of decapping protein 2 (Dcp2) was inversely increased in the above specimens. It showed that increased Dcp2 could promote the degradation of lnc13 by separating the lnc13‐hnRNPD compound and relieve subsequent inflammatory respond after LPS stimulating.117 In the cytoplasm of human coeliac disease patient samples and macrophages, the functional characteristics of increased cardiac and apoptosis‐related lncRNA (Carlr) was defined, which elevated the expression of NF‐κB‐regulated genes through binding to p65 and letting it released from IκBα. Aberrant activation of NF‐κB might cause the overproduction of inflammatory cytokines and mucosal inflammation that contributes to intestinal inflammatory diseases. At the same time, NF‐κB signalling caused changes in other lncRNAs that have been shown to be important for both cytokines production and subsequent resolution of inflammation. Compared with control patients, three of the putative Carlr‐regulated targets, TNFAIP3, IL1B and PTGS2, showed statistically significant higher expression in coeliac patients.118
5.4. Mycobacterial infections
Macrophages can phagocytose invading mycobacterium tuberculosis (Mtb), so it is considered as the main residence for Mtb and gives the ability to escape damage. LncRNAs have proved to involve in anti‐mycobacterial infection, but their effects on this response remain unelucidated.119 An arrayed analysis of lncRNAs was conducted in human macrophages infected with M. bovis Calmette‐Guerin (BCG), and showed that several lncRNAs expression including MEG3 was suppressed after infection. MEG3 was associated with mTOR and PI3K/AKT pathway and involved in autophagy regulation of macrophages using pathway analysis. And persistent decline in MEG3 expression was observed in IFN‐γ‐induced autophagy of infected macrophages. Knockdown of MEG3 in macrophages would help to destroy the intracellular M. bovis BCG of macrophages.120 Another microarray analysis conducted in human macrophages with H37Ra or H37Rv about 72 hours, revealed that lncRNAs diversity expressed in macrophages. In particular, up‐regulated MIR3945HG V1 and MIR3945HG V2 showed potential diagnostic markers for Mtb infection, but their functions were still unknown.121
5.5. Other relevant diseases
Silicosis is one of the most severe occupational diseases, and shows common histological changes in persistent inflammation and pulmonary fibrosis. Studies have indicated that silica particles could promote the liberation of copious oxidants and inflammatory factors in macrophages and epithelial cells, which caused a deposition in extracellular matrix, and finally led to pulmonary fibrosis.122 Silica‐induced pulmonary fibrosis, macrophages exposed to silica or fibroblasts exposed to TGF‐β were decreased the miR‐489 level.123 MiR‐489 possesses the anti‐fibrosis action via decreasing MyD88 and Smad3 target. LncRNA cardiac hypertrophy‐related factor (CHRF) was elevated and accelerated inflammation responses and fibrosis in the silica‐treated RAW264.7, then acted as a sponge of miR‐489, reversing the inhibitory effect of miR‐489124 (Figure 4).
Figure 4.
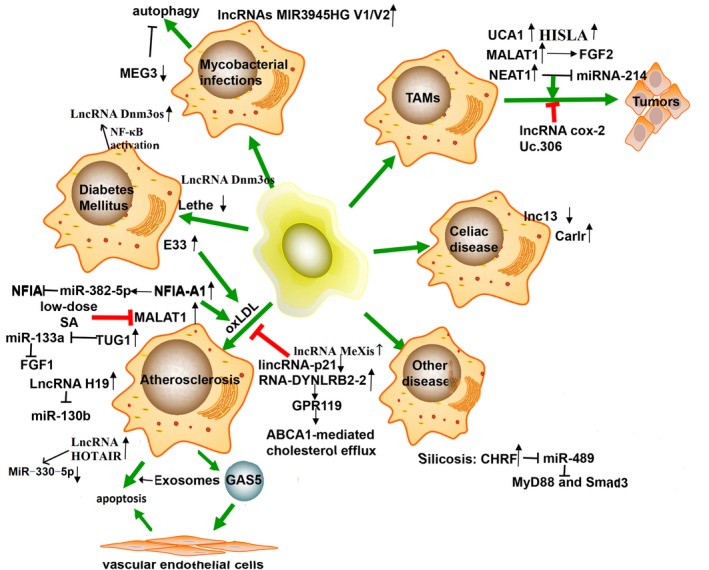
Dysregulation of lncRNAs and macrophages in inflammatory diseases. Macrophages are closely related to a number of physiological courses and pathological changes. Major lncRNAs involved in each macrophage‐mediated key processes have been presented. TAMs: tumour‐associated macrophages
6. CONCLUSION
Macrophages are remarkably versatile, and can elicite extensive responses to environmental stimulus while maintaining organ homeostasis and preventing autoimmunity. LncRNAs have been demonstrated to participate in multiple stages of immune cell development and in pathogen responses. Combining currently published articles, we witness an significant growth in the understanding of lncRNAs regulation on macrophage biology, including its development, differentiation and activation. Such understandings of how lncRNAs function in macrophages may influence future therapies for inflammation‐associated diseases, and add a new perspective for the innate inflammatory response. The same lncRNA expressed in macrophages can play diversities of roles in different physiological and pathological processes, such as lncRNA MALAT1. Some lncRNAs, however, both inhibit and promote inflammation reponses through different mechanisms in macrophages in the case of internal environment disorder, like linc RNA‐Cox2. Besides, this review summarizes the interaction between lncRNAs and some miRNAs in the process of macrophage development. Exosomal‐derived lncRNAs also are involved in diverse biological progress.125 Macrophage‐secreted exosomes can transport lncRNAs, regulate and modify its development and polarization, indicating its remote regulation in life processes, and adding a new dimension to macrophage functions. Circular RNAs (circRNAs) are new molecules in the regulation of post‐transcriptional genes expression and important to the pathological process of several diseases. According to previous researches, circRNAs play a crucial role in fine‐tuning the level of miRNA‐mediated regulation of gene expression by sequestering miRNAs. However, the effects of circRNAs in macrophage differentiation and polarization have not to be widely explored. Whether circRNA involved in the activation of macrophages and whether it affect the development of macrophages by regulating lncRNAs require more further experiments to detect.126 Unfortunately, there is a defect in our paper that many studies on the mechanisms of lncRNAs expressed in macrophages have not been studied intensively. It is expected that more exploration about lncRNAs in regulating innate immune response and macrophage‐associated biological processes will be carried out. In conclusion, lncRNAs harmonizes in the development and function of macrophages, which can prevent the human body from being damaged by internal and external stimuli without generating an excessive immune response. Increased or depressed lncRNAs in macrophages might be regarded as potential therapeutic targets and diagnostic biomarkers, and provided a new sight into the treatment of inflammatory diseases and cancers.
CONFLICT OF INTEREST
The authors confirm that there are no conflicts of interest.
AUTHORS CONTRIBUTION
Yixn Xie and Min Wang initiated this review, read lots of literature and wrote the manuscript. Other authors revised our first draft and provided valuable comments. All authors read the manuscript and approved it.
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China (81470133) and the Fundamental Research Funds for the Central Universities of Central South University (2017zzts896, 2017zzts844).
Xie Y, Wang M, Tian J, et al. Long non‐coding RNA expressed in macrophage co‐varies with the inflammatory phenotype during macrophage development and polarization. J Cell Mol Med. 2019;23:6530–6542. 10.1111/jcmm.14557
REFERENCES
- 1. Locati M, Mantovani A, Sica A. Macrophage activation and polarization as an adaptive component of innate immunity. Adv Immunol. 2013;120:163‐184. [DOI] [PubMed] [Google Scholar]
- 2. Verbist KC, Wang R, Green DR. T cell metabolism and the immune response. Semin Immunol. 2012;24:399‐404. [DOI] [PubMed] [Google Scholar]
- 3. Huang J‐K, Ma L, Song W‐H, et al. LncRNA‐MALAT1 promotes angiogenesis of thyroid cancer by modulating tumor‐associated macrophage FGF2 protein secretion. J Cell Biochem. 2017;118:4821‐4830. [DOI] [PubMed] [Google Scholar]
- 4. Beirão B, Raposo T, Pang LY, Argyle DJ. Canine mammary cancer cells direct macrophages toward an intermediate activation state between M1/M2. BMC Vet Res. 2015;11:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weisser SB, McLarren KW, Kuroda E, et al. Generation and characterization of murine alternatively activated macrophages. Methods Mol Biol. 2013;946:225‐239. [DOI] [PubMed] [Google Scholar]
- 6. Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11:889‐896. [DOI] [PubMed] [Google Scholar]
- 7. Murray P, Allen J, Biswas S, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou D, Huang C, Lin Z, et al. Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways. Cell Signal. 2014;26:192‐197. [DOI] [PubMed] [Google Scholar]
- 9. Lu J, Xie L, Liu C, Zhang Q, Sun S. PTEN/PI3k/AKT regulates macrophage polarization in emphysematous mice. Scand J Immunol. 2017;85:395‐405. [DOI] [PubMed] [Google Scholar]
- 10. Zhou L, Zhuo H, Ouyang H, et al. Glycoprotein non‐metastatic melanoma protein b (Gpnmb) is highly expressed in macrophages of acute injured kidney and promotes M2 macrophages polarization. Cell Immunol. 2017;316:53‐60. [DOI] [PubMed] [Google Scholar]
- 11. Yang M, Liu J, Piao C, Shao J, Du J. ICAM‐1 suppresses tumor metastasis by inhibiting macrophage M2 polarization through blockade of efferocytosis. Cell Death Dis. 2015;6:e1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wan J, Benkdane M, Teixeira‐Clerc F, et al. M2 Kupffer cells promote M1 Kupffer cell apoptosis: a protective mechanism against alcoholic and nonalcoholic fatty liver disease. Hepatology. 2014;59:130‐142. [DOI] [PubMed] [Google Scholar]
- 13. Mantovani A, Biswas SK, Galdiero MR, et al. Macrophage plasticity and polarization in tissue repair and remodelling. J Pathol. 2013;229:176‐185. [DOI] [PubMed] [Google Scholar]
- 14. Beermann J, Piccoli M‐T, Viereck J, Thum T. Non‐coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297‐1325. [DOI] [PubMed] [Google Scholar]
- 15. Jiang Q, Ma R, Wang J, et al. LncRNA2Function: a comprehensive resource for functional investigation of human lncRNAs based on RNA‐seq data. BMC Genom. 2015;16(Suppl 3):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xie ZC, Dang YW, Wei DM, et al. Clinical significance and prospective molecular mechanism of MALAT1 in pancreatic cancer exploration: a comprehensive study based on the GeneChip, GEO, Oncomine, and TCGA databases. Onco Targets Ther. 2017;10:3991‐4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu SY, Tang L, Zhou SH. Long noncoding RNAs: new players in ischaemia‐reperfusion injury. Heart Lung Circ. 2018;27:322‐332. [DOI] [PubMed] [Google Scholar]
- 18. Li J, Tian H, Yang J, Gong Z. Long noncoding RNAs regulate cell growth, proliferation, and apoptosis. Dna Cell Biol. 2016;35:459‐470. [DOI] [PubMed] [Google Scholar]
- 19. Atianand MK, Caffrey DR, Fitzgerald KA. Immunobiology of long noncoding RNAs. Annu Rev Immunol. 2017;35:177‐198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stathopoulou C, Kapsetaki M, Stratigi K, et al. Long non‐coding RNA SeT and miR‐155 regulate the Tnfalpha gene allelic expression profile. PLoS ONE. 2017;12:e184788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mathy NW, Chen X. Long non‐coding RNAs (lncRNAs) and their transcriptional control of inflammatory responses. J Biol Chem. 2017;292:12375‐12382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gezer U, Özgür E, Cetinkaya M, Isin M, Dalay N. Long non‐coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol Int. 2014;38:1076‐1079. [DOI] [PubMed] [Google Scholar]
- 23. Li Z, Dou P, Liu T, He S. Application of long noncoding RNAs in osteosarcoma: biomarkers and therapeutic targets. Cell Physiol Biochem. 2017;42:1407‐1419. [DOI] [PubMed] [Google Scholar]
- 24. Guttman M, Rinn JL. Modular regulatory principles of large non‐coding RNAs. Nature. 2012;482:339‐346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fatica A, Bozzoni I. Long non‐coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7‐21. [DOI] [PubMed] [Google Scholar]
- 26. Quinn JJ, Chang HY. Unique features of long non‐coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47‐62. [DOI] [PubMed] [Google Scholar]
- 27. Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Novikova IV, Hennelly SP, Tung C‐S, Sanbonmatsu KY. Rise of the RNA machines: exploring the structure of long non‐coding RNAs. J Mol Biol. 2013;425:3731‐3746. [DOI] [PubMed] [Google Scholar]
- 29. Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300‐307. [DOI] [PubMed] [Google Scholar]
- 30. Managadze D, Rogozin IB, Chernikova D, Shabalina SA, Koonin EV. Negative correlation between expression level and evolutionary rate of long intergenic noncoding RNAs. Genome Biol Evol. 2011;3:1390‐1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775‐1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2008;105:716‐721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Geisler S, Coller J. RNA in unexpected places: long non‐coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699‐712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee JT, Bartolomei MS. X‐inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308‐1323. [DOI] [PubMed] [Google Scholar]
- 35. Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311‐1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ørom UA, Shiekhattar R. Long noncoding RNAs usher in a new era in the biology of enhancers. Cell. 2013;154:1190‐1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gong C, Maquat LE. lncRNAs transactivate STAU1‐mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature. 2011;470:284‐288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yoon J‐H, Abdelmohsen K, Srikantan S, et al. LincRNA‐p21 suppresses target mRNA translation. Mol Cell. 2012;47:648‐655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Karreth FA, Tay Y, Perna D, et al. In vivo identification of tumor‐ suppressive PTEN ceRNAs in an oncogenic BRAF‐induced mouse model of melanoma. Cell. 2011;147:382‐395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lin C, Yang L. Long noncoding RNA in cancer: wiring signaling circuitry. Trends Cell Biol. 2018;28(4):287‐301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barile L, Vassalli G. Exosomes: therapy delivery tools and biomarkers of diseases. Pharmacol Ther. 2017;174:63‐78. [DOI] [PubMed] [Google Scholar]
- 42. Shi C, Pamer EG. Monocyte recruitment during infection and inflammation. Nat Rev Immunol. 2011;11:762‐774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene. 2007;26:6816‐6828. [DOI] [PubMed] [Google Scholar]
- 44. Roy S. miRNA in macrophage development and function. Antioxid Redox Signal. 2016;25:795‐804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen M, Lin H, Shen C, et al. PU.1‐regulated long noncoding RNA Lnc‐MC controls human monocyte/macrophage differentiation through interaction with MicroRNA‐199a‐5p. Mol Cell Biol. 2015;35:415‐429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lin HS, Gong JN, Su R, et al. miR‐199a‐5p inhibits monocyte/macrophage differentiation by targeting the activin A type 1B receptor gene and finally reducing C/EBPalpha expression. J Leukoc Biol. 2014;96:1023‐1035. [DOI] [PubMed] [Google Scholar]
- 47. Chávez‐Galán L, Olleros ML, Vesin D, et al. Much more than M1 and M2 macrophages, there are also CD169(+) and TCR(+) macrophages. Front Immunol. 2015;6:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang CA, Li JP, Yen JC, et al. lncRNA NTT/PBOV1 axis promotes monocyte differentiation and is elevated in rheumatoid arthritis. Int J Mol Sci. 2018;19:2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mao X, Su Z, Mookhtiar AK. Long non‐coding RNA: a versatile regulator of the nuclear factor‐kappaB signalling circuit. Immunology. 2017;150:379‐388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Murphy MB, Medvedev AE. Long noncoding RNAs as regulators of Toll‐like receptor signaling and innate immunity. J Leukoc Biol. 2016;99:839‐850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF‐kappaB regulatory pathways. Annu Rev Biochem. 2009;78:769‐796. [DOI] [PubMed] [Google Scholar]
- 52. He X, Li Y, Li C, et al. USP2a negatively regulates IL‐1beta‐ and virus‐induced NF‐kappaB activation by deubiquitinating TRAF6. J Mol Cell Biol. 2013;5:39‐47. [DOI] [PubMed] [Google Scholar]
- 53. Du M, Yuan L, Tan X, et al. The LPS‐inducible lncRNA Mirt2 is a negative regulator of inflammation. Nat Commun. 2017;8:2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kawai T, Akira S. Toll‐like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637‐650. [DOI] [PubMed] [Google Scholar]
- 55. Kagan JC, Su T, Horng T, et al. TRAM couples endocytosis of Toll‐like receptor 4 to the induction of interferon‐beta. Nat Immunol. 2008;9:361‐368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ye Y, Xu Y, Lai YU, et al. Long non‐coding RNA cox‐2 prevents immune evasion and metastasis of hepatocellular carcinoma by altering M1/M2 macrophage polarization. J Cell Biochem. 2018;119:2951‐2963. [DOI] [PubMed] [Google Scholar]
- 57. Mao AP, Shen J, Zuo Z. Expression and regulation of long noncoding RNAs in TLR4 signaling in mouse macrophages. BMC Genom. 2015;16:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chan J, Atianand M, Jiang Z, et al. Cutting edge: a natural antisense transcript, AS‐IL1α, controls inducible transcription of the proinflammatory cytokine IL‐1α. J Immunol. 2015;195:1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Krawczyk M, Emerson BM. p50‐associated COX‐2 extragenic RNA (PACER) activates COX‐2 gene expression by occluding repressive NF‐kappaB complexes. eLife. 2014;3:e1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ilott NE, Heward JA, Roux B, et al. Long non‐coding RNAs and enhancer RNAs regulate the lipopolysaccharide‐induced inflammatory response in human monocytes. Nat Commun. 2014;5:3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Anggayasti WL, Mancera RL, Bottomley S, et al. The self‐association of HMGB1 and its possible role in the binding to DNA and cell membrane receptors. FEBS Lett. 2017;591:282‐294. [DOI] [PubMed] [Google Scholar]
- 62. Ma S, Ming Z, Gong A‐Y, et al. A long noncoding RNA, lincRNA‐Tnfaip3, acts as a coregulator of NF‐κB to modulate inflammatory gene transcription in mouse macrophages. FASEB J. 2017;31:1215‐1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yang H, Bi Y, Xue L, et al. Multifaceted modulation of SIRT1 in cancer and inflammation. Crit Rev Oncog. 2015;20:49‐64. [DOI] [PubMed] [Google Scholar]
- 64. Jia Y, Li Z, Cai W, et al. SIRT1 regulates inflammation response of macrophages in sepsis mediated by long noncoding RNA. Biochim Biophys Acta. 2017;1864:784‐792. [DOI] [PubMed] [Google Scholar]
- 65. Lu Y, Liu X, Xie M, et al. The NF‐kappaB‐responsive long noncoding RNA FIRRE regulates posttranscriptional regulation of inflammatory gene expression through interacting with hnRNPU. J Immunol. 2017;199:3571‐3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang W, Lou C, Gao J, Zhang X, Du Y. LncRNA SNHG16 reverses the effects of miR‐15a/16 on LPS‐induced inflammatory pathway. Biomed Pharmacother. 2018;106:1661‐1667. [DOI] [PubMed] [Google Scholar]
- 67. Carpenter S, Aiello D, Atianand MK, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341:789‐792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hu G, Gong A‐Y, Wang Y, et al. LincRNA‐Cox2 promotes late inflammatory gene transcription in macrophages through modulating SWI/SNF‐mediated chromatin remodeling. J Immunol. 2016;196:2799‐2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Covarrubias S, Robinson EK, Shapleigh B, et al. CRISPR/Cas‐based screening of long non‐coding RNAs (lncRNAs) in macrophages with an NF‐kappaB reporter. J Biol Chem. 2017;292:20911‐20920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tong Q, Gong AY, Zhang XT, et al. LincRNA‐Cox2 modulates TNF‐alpha‐induced transcription of Il12b gene in intestinal epithelial cells through regulation of Mi‐2/NuRD‐mediated epigenetic histone modifications. FASEB J. 2016;30:1187‐1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zgheib C, Hodges MM, Hu J, Liechty KW, Xu J. Long non‐coding RNA Lethe regulates hyperglycemia‐induced reactive oxygen species production in macrophages. PLoS ONE. 2017;12:e177453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Spurlock CR, Tossberg JT, Matlock BK, et al. Methotrexate inhibits NF‐kappaB activity via long intergenic (noncoding) RNA‐p21 induction. Arthritis Rheumatol. 2014;66:2947‐2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cui H, Xie NA, Tan Z, et al. The human long noncoding RNA lnc‐IL7R regulates the inflammatory response. Eur J Immunol. 2014;44:2085‐2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Li Z, Chao TC, Chang KY, et al. The long noncoding RNA THRIL regulates TNF expression through its interaction with hnRNPL. Proc Natl Acad Sci USA. 2014;111:1002‐1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Morris MC, Gilliam EA, Li L. Innate immune programing by endotoxin and its pathological consequences. Front Immunol. 2014;5:680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Stratigi K, Kapsetaki M, Aivaliotis M, Town T, Flavell RA, Spilianakis CG. Spatial proximity of homologous alleles and long noncoding RNAs regulate a switch in allelic gene expression. Proc Natl Acad Sci U S A. 2015;112:E1577‐E1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Liu B, Sun L, Liu Q, et al. A cytoplasmic NF‐kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370‐381. [DOI] [PubMed] [Google Scholar]
- 78. Atianand MK, Hu W, Satpathy AT, et al. A long noncoding RNA lincRNA‐EPS acts as a transcriptional brake to restrain inflammation. Cell. 2016;165:1672‐1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhao G, Su Z, Song D, Mao Y, Mao X. The long noncoding RNA MALAT1 regulates the lipopolysaccharide‐induced inflammatory response through its interaction with NF‐κB. FEBS Lett. 2016;17:2884‐2895. [DOI] [PubMed] [Google Scholar]
- 80. Dai L, Zhang G, Cheng Z, et al. Knockdown of LncRNA MALAT1 contributes to the suppression of inflammatory responses by up‐regulating miR‐146a in LPS‐induced acute lung injury. Connect Tissue Res. 2018;59:581‐592. [DOI] [PubMed] [Google Scholar]
- 81. Zhang H, Xue C, Wang Y, et al. Deep RNA sequencing uncovers a repertoire of human macrophage long intergenic noncoding RNAs modulated by macrophage activation and associated with cardiometabolic diseases. J Am Heart Assoc. 2017;6:e007431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Porta C, Rimoldi M, Raes G, et al. Tolerance and M2 (alternative) macrophage polarization are related processes orchestrated by p50 nuclear factor kappaB. Proc Natl Acad Sci USA. 2009;106:14978‐14983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sun Y, Jin X, Liu X, et al. MicroRNA let‐7i regulates dendritic cells maturation targeting interleukin‐10 via the Janus kinase 1‐signal transducer and activator of transcription 3 signal pathway subsequently induces prolonged cardiac allograft survival in rats. J Heart Lung Transplant. 2016;35:378‐388. [DOI] [PubMed] [Google Scholar]
- 84. Gao X, Ge J, Li W, Zhou W, Xu L. LncRNA KCNQ1OT1 ameliorates particle‐induced osteolysis through inducing macrophage polarization by inhibiting miR‐21a‐5p. Biol Chem. 2017;399:375‐386. [DOI] [PubMed] [Google Scholar]
- 85. Huang Z, Luo Q, Yao F, et al. Identification of differentially expressed long non‐coding RNAs in polarized macrophages. Sci Rep. 2016;6:19705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Cao JI, Dong R, Jiang LI, et al. LncRNA‐MM2P identified as a modulator of macrophage M2 polarization. Cancer Immunol Res. 2018. [DOI] [PubMed] [Google Scholar]
- 87. Chi X, Ding B, Zhang L, Zhang J, Wang J, Zhang W. lncRNA GAS5 promotes M1 macrophage polarization via miR‐455‐5p/SOCS3 pathway in childhood pneumonia. J Cell Physiol. 2018;234(8):13242‐13251. [DOI] [PubMed] [Google Scholar]
- 88. Khan MA, Assiri AM, Broering DC. Complement and macrophage crosstalk during process of angiogenesis in tumor progression. J Biomed Sci. 2015;22:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Li J‐H, Zhang S‐Q, Qiu X‐G, Zhang S‐J, Zheng S‐H, Zhang D‐H. Long non‐coding RNA NEAT1 promotes malignant progression of thyroid carcinoma by regulating miRNA‐214. Int J Oncol. 2017;50:708‐716. [DOI] [PubMed] [Google Scholar]
- 90. Wu Q, Wu X, Ying X, et al. Suppression of endothelial cell migration by tumor associated macrophage‐derived exosomes is reversed by epithelial ovarian cancer exosomal lncRNA. Cancer Cell Int. 2017;17:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Luo H‐L, Chen J, Luo T, et al. Downregulation of macrophage‐derived T‐UCR uc.306 associates with poor prognosis in hepatocellular carcinoma. Cell Physiol Biochem. 2017;42:1526‐1539. [DOI] [PubMed] [Google Scholar]
- 92. Chen S, Shao C, Xu M, et al. Macrophage infiltration promotes invasiveness of breast cancer cells via activating long non‐coding RNA UCA1. Int J Clin Exp Pathol. 2015;8:9052‐9061. [PMC free article] [PubMed] [Google Scholar]
- 93. Chen F, Chen J, Yang L, et al. Extracellular vesicle‐packaged HIF‐1α‐stabilizing lncRNA from tumour‐associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol. 2019;21(4):498‐510. [DOI] [PubMed] [Google Scholar]
- 94. Liu J, Ding D, Jiang Z, et al. Long non‐coding RNA CCAT1/miR‐148a/PKCζ prevents cell migration of prostate cancer by altering macrophage polarization. Prostate. 2019;79(1):105‐112. [DOI] [PubMed] [Google Scholar]
- 95. Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473:317‐325. [DOI] [PubMed] [Google Scholar]
- 96. Kunjathoor VV, Febbraio M, Podrez EA, et al. Scavenger receptors class A‐I/II and CD36 are the principal receptors responsible for the uptake of modified low density lipoprotein leading to lipid loading in macrophages. J Biol Chem. 2002;277:49982‐49988. [DOI] [PubMed] [Google Scholar]
- 97. Huangfu N, Xu Z, Zheng W, et al. LncRNA MALAT1 regulates oxLDL‐induced CD36 expression via activating beta‐catenin. Biochem Biophys Res Commun. 2018;495:2111‐2117. [DOI] [PubMed] [Google Scholar]
- 98. Gast M, Rauch BH, Nakagawa S, et al. Immune system‐mediated atherosclerosis caused by deficiency of long noncoding RNA MALAT1 in ApoE‐/‐ mice. Cardiovasc Res. 2018;115:302‐314. [DOI] [PubMed] [Google Scholar]
- 99. Han Y, Qiu H, Pei X, et al. Low‐dose sinapic acid abates the pyroptosis of macrophages via downregulation of lncRNA‐MALAT1 in rats with diabetic atherosclerosis. J Cardiovasc Pharmacol. 2018;2:104‐112. [DOI] [PubMed] [Google Scholar]
- 100. Liu J, Huang GQ, Ke ZP, et al. Silence of long intergenic noncoding RNA HOTAIR ameliorates oxidative stress and inflammation response in ox‐LDL‐treated human macrophages by upregulating miR‐330‐5p. J Cell Physiol. 2018;234(4):5134‐5142. [DOI] [PubMed] [Google Scholar]
- 101. Wu G, Cai J, Han YU, et al. LincRNA‐p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation. 2014;130:1452‐1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Zhang L, Cheng H, Yue Y, Li S, Zhang D, He R. TUG1 knockdown ameliorates atherosclerosis via up‐regulating the expression of miR‐133a target gene FGF1. Cardiovasc Pathol. 2017;33:6‐15. [DOI] [PubMed] [Google Scholar]
- 103. Han Y, Ma J, Wang J, Wang L. Silencing of H19 inhibits the adipogenesis and inflammation response in ox‐LDL‐treated Raw264.7 cells by up‐regulating miR‐130b. Mol Immunol. 2018;93:107‐114. [DOI] [PubMed] [Google Scholar]
- 104. Hu YW, Zhao JY, Li SF, et al. RP5‐833A20.1/miR‐382‐5p/NFIA‐dependent signal transduction pathway contributes to the regulation of cholesterol homeostasis and inflammatory reaction. Arterioscler Thromb Vasc Biol. 2014;35:87‐101. [DOI] [PubMed] [Google Scholar]
- 105. Holdt LM, Teupser D. Long noncoding RNA‐MicroRNA pathway controlling nuclear factor IA, a novel atherosclerosis modifier gene. Arterioscler Thromb Vasc Biol. 2015;35:7‐8. [DOI] [PubMed] [Google Scholar]
- 106. Ye D, Lammers B, Zhao Y, et al. ATP‐binding cassette transporters A1 and G1, HDL metabolism, cholesterol efflux, and inflammation: important targets for the treatment of atherosclerosis. Curr Drug Targets. 2011;12:647‐660. [DOI] [PubMed] [Google Scholar]
- 107. Hu Y‐W, Yang J‐Y, Ma X, et al. A lincRNA‐DYNLRB2‐2/GPR119/GLP‐1R/ABCA1‐dependent signal transduction pathway is essential for the regulation of cholesterol homeostasis. J Lipid Res. 2014;55:681‐697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Sallam T, Jones M, Thomas BJ, et al. Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA. Nat Med. 2018;24:304‐312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Chen L, Yao H, Hui J‐Y, et al. Global transcriptomic study of atherosclerosis development in rats. Gene. 2016;592:43‐48. [DOI] [PubMed] [Google Scholar]
- 110. Krell J, Frampton AE, Mirnezami R, et al. Growth arrest‐specific transcript 5 associated snoRNA levels are related to p53 expression and DNA damage in colorectal cancer. PLoS ONE. 2014;9:e98561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Ye J, Wang C, Wang D, Yuan H. LncRBA GSA5, up‐regulated by ox‐LDL, aggravates inflammatory response and MMP expression in THP‐1 macrophages by acting like a sponge for miR‐221. Exp Cell Res. 2018;369:348‐355. [DOI] [PubMed] [Google Scholar]
- 112. Chen L, Yang W, Guo Y, et al. Exosomal lncRNA GAS5 regulates the apoptosis of macrophages and vascular endothelial cells in atherosclerosis. PLoS ONE. 2017;12:e185406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Das S, Reddy MA, Senapati P, et al. Diabetes mellitus‐induced long noncoding RNA Dnm3os regulates macrophage functions and inflammation via nuclear mechanisms. Arterioscler Thromb Vasc Biol. 2018;38:1806‐1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Reddy MA, Chen Z, Park JT, et al. Regulation of inflammatory phenotype in macrophages by a diabetes‐induced long noncoding RNA. Diabetes. 2014;63:4249‐4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge R‐M, Chang HY. A mammalian pseudogene lncRNA at the interface of inflammation and anti‐inflammatory therapeutics. eLife. 2013;2:e762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Fernandez‐Jimenez N, Castellanos‐Rubio A, Plaza‐Izurieta L, et al. Coregulation and modulation of NFkappaB‐related genes in celiac disease: uncovered aspects of gut mucosal inflammation. Hum Mol Genet. 2014;23:1298‐1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Castellanos‐Rubio A, Fernandez‐Jimenez N, Kratchmarov R, et al. A long noncoding RNA associated with susceptibility to celiac disease. Science. 2016;352:91‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Castellanos‐Rubio A, Kratchmarov R, Sebastian M, et al. Cytoplasmic form of Carlr lncRNA facilitates inflammatory gene expression upon NF‐kappaB activation. J Immunol. 2017;199:581‐588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Aune TM, Spurlock CR. Long non‐coding RNAs in innate and adaptive immunity. Virus Res. 2016;212:146‐160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Pawar K, Hanisch C, Palma Vera SE, Einspanier R, Sharbati S. Down regulated lncRNA MEG3 eliminates mycobacteria in macrophages via autophagy. Sci Rep. 2016;6:19416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Yang X, Yang J, Wang J, et al. Microarray analysis of long noncoding RNA and mRNA expression profiles in human macrophages infected with Mycobacterium tuberculosis. Sci Rep. 2016;6:38963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Beamer CA, Migliaccio CT, Jessop F, Trapkus M, Yuan D, Holian A. Innate immune processes are sufficient for driving silicosis in mice. J Leukoc Biol. 2010;88:547‐557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Ji X, Wu B, Fan J, et al. The anti‐fibrotic effects and mechanisms of MicroRNA‐486‐5p in pulmonary fibrosis. Sci Rep. 2015;5:14131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Wu Q, Han L, Yan W, et al. miR‐489 inhibits silica‐induced pulmonary fibrosis by targeting MyD88 and Smad3 and is negatively regulated by lncRNA CHRF. Sci Rep. 2016;6:30921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Zhang W, Jiang X, Bao J, Wang YI, Liu H, Tang L. Exosomes in pathogen infections: a bridge to deliver molecules and link functions. Front Immunol. 2018;9:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Chen X, Ouyang Z, Shen YI, et al. CircRNA_28313/miR‐195a/CSF1 axis modulates osteoclast differentiation to affect OVX‐induced bone absorption in mice. RNA Biol. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]


