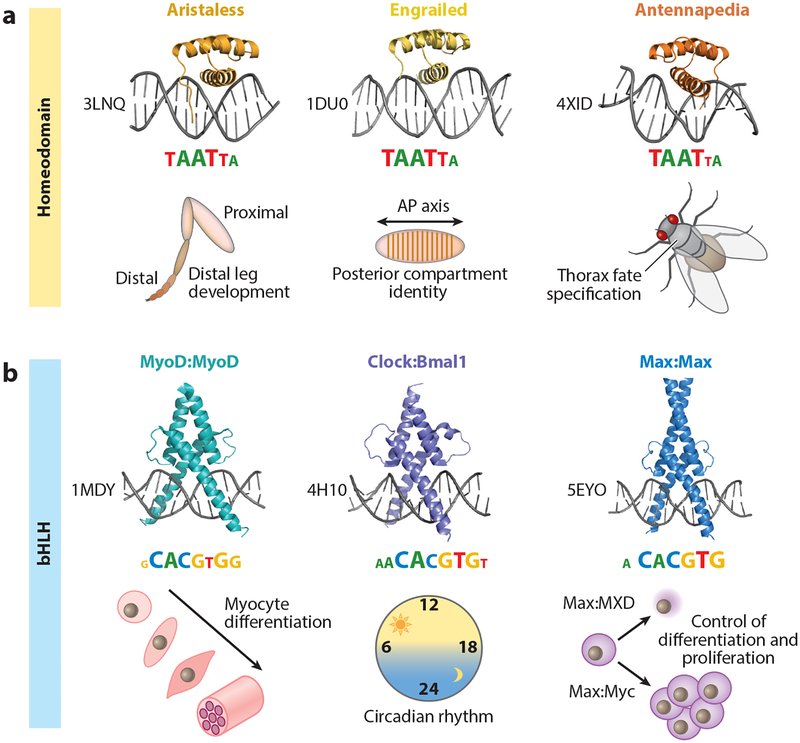
Figure 1.
Shared binding mechanisms and sequence recognition among transcription factor (TF) paralogs with distinct in vivo functions. (a) Crystal structures, binding motifs, and broad in vivo functions of three examples from the Drosophila melanogaster homeodomain TF family: All three TFs—Aristaless (PDB ID: 3LNQ), Engrailed (PDB ID: 1DU0), and Antennapedia (PDB ID: 4XID)—share a conserved structural fold and recognize similar binding motifs, represented as information content logos (motif logos derived from Noyes et al. 2008). Despite the similarities in DNA recognition and sequence readout, Aristaless, Engrailed, and Antennapedia have distinct in vivo functions: distal leg development (Campbell & Tomlinson 1998), posterior compartment identity along the anterior-posterior (AP) axis (Nusslein-Volhard & Wieschaus 1980), and thoracic fate specification (Struhl 1982), respectively. (b) Crystal structures, binding motifs, and broad in vivo functions of three homo- and heterodimeric examples from the Mus musculus and Homo sapiens basic helix-loop-helix (bHLH) TF family: MyoD:MyoD (PDB ID: 1MDY), Clock:Bmal1 (PDB ID: 4H10), and Max:Max (PDB ID: 5EYO). Each of these TF dimers recognizes a highly similar, reverse-complement-symmetric motif [consensus = CACGTG; motifs taken from Jaspar (Khan et al. 2018)], yet has a distinct in vivo function: MyoD is a master regulator of myocyte fate specification (Tapscott et al. 1988); Clock:Bmal1 are central players in establishing a functional circadian clock (Dierickx et al. 2018); and Max is involved in either cell proliferation or differentiation, depending on its binding partners (e.g., Myc or TFs from the MXD class of bHLH TFs) (Carroll et al. 2018).