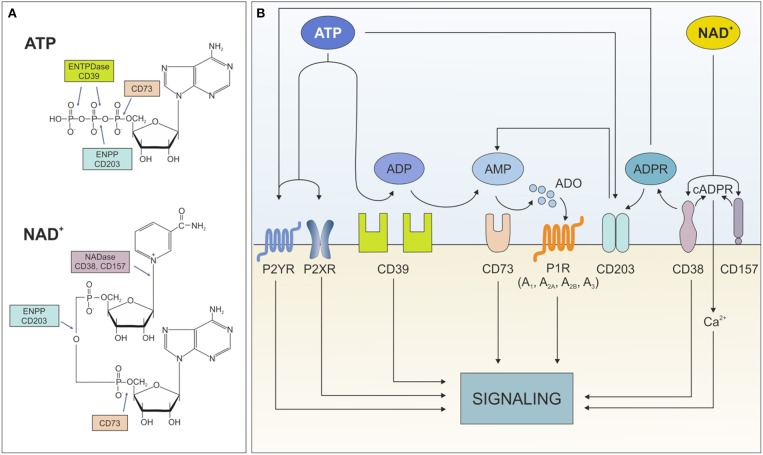
Figure 1.
Ectonucleotidases network. (A) Structure of ATP and NAD+ with indication of cleavage sites by the different ectonucleotidases. (B) Schematic representation of the fate of extracellular ATP and NAD+ dismantling operated by the different ectonucleotidases and the functional consequences of the generated metabolites. ENTPDase, ecto-nucleoside triphosphate diphosphohydrolase; ENPP, ecto-phosphodiesterase/nucleotide phosphohydrolase; ATP, adenine-triphosphate; ADP, adenine-diphosphate; AMP, adenine-monophosphate; ADO, adenosine; P2XR, ATP-gated P2X receptor cation channel; P2YR, G protein-coupled P2Y receptor; P1R, adenosine purinergic receptor; NAD, nicotinamide adenine dinucleotide; ADPR, ADP-ribose; cADPR, cyclic-ADPR; Ca2+, calcium ions.