Figure 1. Pairwise energy functions.
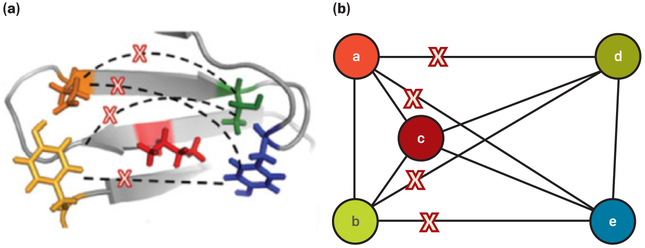
(a) Pairwise energy functions compute energies between pairs of mutable residues (colored) in a protein design problem, but in practice many pairs have very small interaction energies (marked with Xs).
(b) A sparse residue interaction graph (SPRIG) has mutable residues as nodes; edges with small interaction energies can be deleted, enabling highly efficient protein design computations. Figure adapted with permission from Jou et al.20
