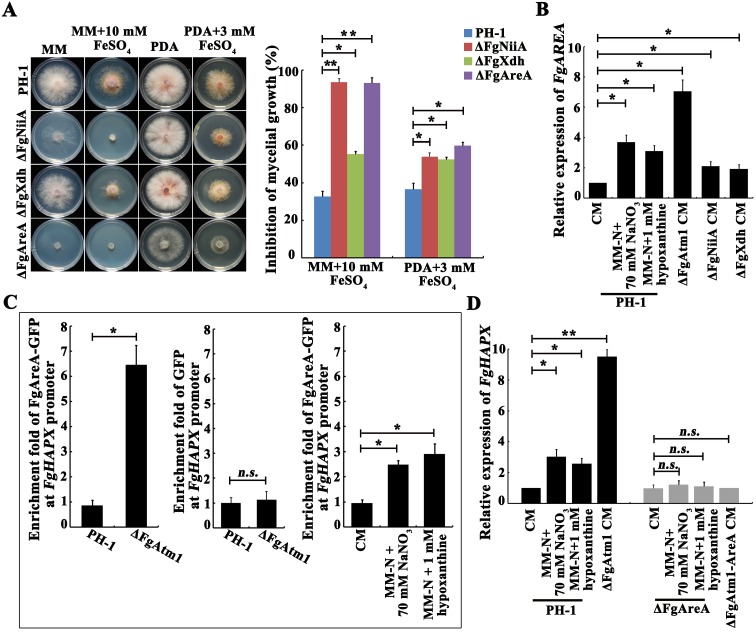
Fig 3. The reduced activity of cytoplasmic Fe-S proteins activates transcription of the FgAreA-HapX cascade.
(A) Sensitivity of the wild type, ΔFgNiiA, ΔFgXdh and ΔFgAreA to FeSO4. A 5-mm mycelial plug of each strain was inoculated on MM or PDA without or with FeSO4 at the indicated concentration, and then incubated at 25°C for 3 days. Mycelial growth inhibition of each treatment was calculated after a 3-day-incubation. Means and standard errors were calculated from three repeats. Significance was measured using an unpaired t-test (*p < 0.05, **p < 0.01). (B) Relative transcription levels of FgAREA treated by NaNO3 or hypoxanthine, or deletion of FgATM1, FgNIIA or FgXDH. PH-1 was treated with MM-N+70 mM NaNO3 or MM-N+1 mM hypoxanthine for 4 hours after culture in CM at 25°C for 1 day. The expression level in the wild type without treatment was set as 1. Means and standard errors were calculated from three repeats. Significance was measured using unpaired t-test (*p < 0.05). (C) The enrichment of FgAreA-GFP at the promoter of FgHAPX was induced by the treatment with NaNO3 or hypoxanthine, or deletion of FgATM1. Each strain was treated with MM-N+70 mM NaNO3 or MM-N+1 mM hypoxanthine for 4 hours after culture in CM at 25°C for 1 day. ChIP- and input-DNA samples were quantified by quantitative PCR assay. A control reaction was processed in parallel with rabbit IgG and PH-1 transformed only with GFP used as a negative control for detecting GFP enrichment at the FgHAPX promoter. Means and standard errors were calculated from three repeats. Significance was measured using unpaired t-test (n.s. not significant, *p < 0.05). (D) Relative transcription levels of FgHAPX in the wild-type PH-1 and ΔFgAreA under a non-preferred nitrogen source or in the background of FgATM1 deletion. Each strain was treated with MM-N+70 mM NaNO3 or MM-N+1 mM hypoxanthine for 4 hours after culture in CM at 25°C for 1 day. The expression level in the wild type without treatment was set as 1. Means and standard errors were calculated from three repeats. Significance was measured using unpaired t-test (n.s. not significant, *p < 0.05, **p < 0.01).