Abstract
Purpose
Germline mutations in BRCA1 and BRCA2 confer a significant increase in risk for cancer, and determining pathogenicity of a BRCA variant can guide the clinical management of the disease. About 1/3 of BRCA1 variants reported in the public databases have uncertain clinical significance due to lack of conclusive evidence. This study aims to characterize a novel BRCA1 deletion affecting the + 4 splice donor site identified in an individual with early-onset breast cancer.
Methods
The effect of BRCA1 c.5332+4delA variant on RNA splicing was evaluated by amplifying regions of BRCA1 from cDNA derived from the patient. The proportion of abnormal transcript in the total transcripts was quantified. Loss of heterozygosity (LOH) in tumor tissue was investigated using Sanger sequencing and fragment analysis.
Results
BRCA1 c.5332+4delA caused skipping of exon 21 in patient-derived samples. Semi-quantitative analysis indicated that this aberrant RT-PCR product accounts for about 40% of the total transcript levels. Loss of heterozygosity (LOH) was observed in patient’s tumor tissue.
Conclusions
Our results indicate that the BRCA1 c.5332+4delA variant contributes to cancer predisposition through disruption of normal mRNA splicing. We classify this variant as likely pathogenic.
Keywords: BRCA1, Breast cancer, Germline variant, Splicing, Exon skipping, Loss of heterozygosity, c.5332+4delA, Pathogenic
Introduction
BRCA1 is a major component of the homologous repair (HR) pathway and is responsible for the regulation of DNA double-strand break repair [1, 2]. The BRCA1 protein comprises a highly conserved RING finger domain at the N terminus and two repeats of the BRCA1 C-terminal (BRCT) domain at the C terminus [3]. The BRCT repeats play essential roles in tumor suppression, DNA damage repair, and transcriptional regulation [4]. Deletion of the last 11 amino acids of the second BRCA1 BRCT domain, which disrupts BRCA1 protein folding and restricts its nuclear localization [4, 5], is associated with early-onset breast cancer [6].
Germline mutations in BRCA1 significantly increase lifetime risk of breast and ovarian cancer [7]. By the age of 70 years, the absolute risk of cancer for female BRCA1 mutation carriers is reported to be approximately 60–71% for breast cancer and 39–59% for ovarian cancer [8–11]. However, a substantial proportion of these alterations are classified as variants of uncertain significance (VUS). ClinVar, the freely accessible public archive of genetic variants, has 5622 BRCA1 entries, of which 1863 are VUSs (33%) [12, 13]. The ambiguity of VUS is problematic and complicates cancer risk assessment for patients and their family members who carry these variants of uncertain significance, since they cannot take advantage of cancer prevention measures or therapeutic treatment such as PARP inhibitors.
It is relatively straightforward to interpret germline BRCA truncating mutations and alterations of canonical splice sites that affect the GU–AG rules. Classification of intronic variants beyond the ± 1–2 bps can be challenging because it is not known whether these subtle changes sufficiently and consistently affect normal splicing to predispose to cancer development. Intronic variants could lead to either complete skipping of one or more exons, intronic retention, activation of neighboring cryptic splice sites, or introduction of a new splice site within an exon or intron [14]. In some cases, exon skipping causes a frameshift and generates premature termination codons. The aberrant mRNA is degraded by nonsense-mediated RNA decay, and the abnormal transcript is hard to detect. In other cases, the aberrant splicing products are relatively stable, and an abnormal PCR band is observed after RT-PCR [15]. Alternatively spliced isoforms of BRCA1 occur naturally in non-malignant tissues and several isoforms have been reported to consistently occur in control samples [16, 17]. Comprehensive studies on BRCA1 splicing events in blood-related and healthy breast tissue samples by the ENIGMA consortium, an international network of collaborators focused on determining and disseminating the clinical significance of variants in BRCA1, BRCA2, and other breast cancer-associated genes [18], suggest that non-mutually exclusive splicing events are randomly combined to produce several different naturally occurring BRCA1 isoforms [16], further complicating the interpretation of a potential splicing variant. To date, over 160 pathogenic or likely pathogenic BRCA1 splice site variants are reported in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar).
Several studies have reported mutations occurring at the beginning of the intron 21 of BRCA1 (NM_007294.3, lack of exon 4), including a pathogenic variant c.5332+1G>A that causes aberrant splicing and results in the loss of the entire exon 21 [19–22]. The c.5332+1delG mutation was identified in a Chinese woman with early-onset breast cancer [23] and the mutation c.5332+1G>C was reported in a woman with family history of breast cancer [24]. BRCA1 c.5332G>A and c.5332G>T, which occur at the last nucleotide of exon 21, were shown to similarly affect function in experimental studies and classified as pathogenic variants [25]. In this study, we characterized a VUS affecting the same donor splice site, c.5332+4delA, which was identified through clinical testing. This variant leads to exclusion of the entire exon 21 and presumably causes premature truncation of the protein. Loss of heterozygosity (LOH) analysis in the patient tumor sample demonstrated a strong tendency to induce LOH of the wild-type allele in the tumor. Based on our data, we classify this variant as likely pathogenic.
Materials and methods
The Subject
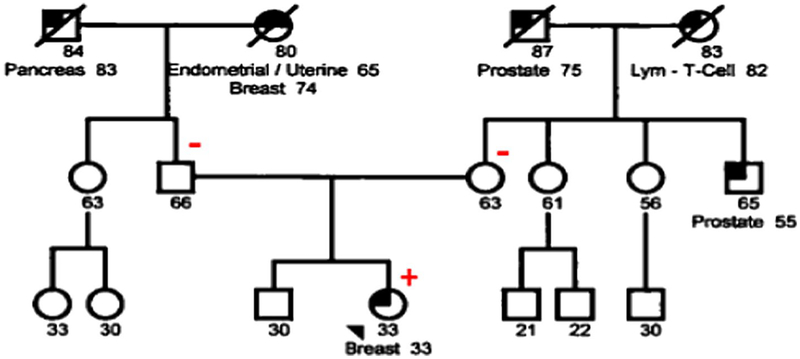
The patient described here is a 33-year-old female who was diagnosed with breast cancer at age 33. Her maternal uncle and grandfather were diagnosed with prostate cancer at 55 and 75 years of age, respectively. Her maternal grandmother was diagnosed with T cell lymphoma in her 80s. The proband’s paternal grandfather was affected with pancreatic cancer at 83. Her paternal grandmother was diagnosed with endometrial cancer at age 65 and breast cancer at 74 (Fig. 1). The BRCA1 c.5332+4delA variant, reported as VUS, was identified through clinical breast cancer panel testing with seven genes evaluated (ATM, BRCA1, BRCA2, CHEK2, PALB2, PTEN, and TP53) in a commercial laboratory. No other reportable variants were identified by sequencing or deletion/duplication in the remaining 6 genes analyzed. Given the uncertain clinical significance of this variant, the patient provided written informed consent for genetic testing as part of a study approved by the Institutional.
Fig. 1.
Patient pedigree. The patient described here is a 33-year-old female who was diagnosed with breast cancer at age 33. Plus and minus signs represent family members with and without the BRCA1 c.5332+4delA variant
Review Board of MSKCC (protocol #96–051 “Clinical Significance of Germline BRCA Mutations”). Peripheral blood samples were collected using the EDTA Blood tube and PAXgene Blood RNA tube (Qiagen, Valencia, CA) and submitted to the Diagnostics Molecular Genetics Laboratory at MSKCC for further analysis. Control RNAs were extracted from unrelated individuals seen at MSKCC who do not carry the BRCA1 variant.
In silico analysis
Sequence data spanning the BRCA1 locus for Homo sapiens [Chromosome 17: 43,044,295–43,125,370 reverse strand] was obtained from the Ensembl Genome Browser (http://www.ensembl.org/index.html).
Primers were designed using the Primer 3 software (http://bioinfo.ut.ee/primer3-0.4.0/). In silico prediction of its effects on splicing was performed using Alamut (Interactive Biosoftware), which includes SSF, MaxEnt, NNSPLICE, GeneSplicer, and HSF tools.
cDNA analysis
Total RNA was extracted using the PAXgene BloodRNA Kit (PreAnalytiX, Qiagen, Valencia, CA) and was subsequently used for cDNA synthesis (SuperScript III First-Strand Synthesis SuperMix, Invitrogen Life Technologies, Carlsbad, CA). Control RNA was extracted from 12 individuals who did not carry the BRCA1 variant. DNase I was used to remove DNA during RNA extraction. Reverse transcription was conducted using SuperScript III First-Strand Synthesis SuperMix (Invitrogen) in the presence of 2.3 μM Oligo(dT)20. PCR was performed using the Jump-Start REDTaq Ready Mix (Sigma), with control cDNA or the patient’s cDNA in the presence of M13-tagged forward and reverse primers (Forward, 5′-GTAAAACGACGGCCA GTA AAG AAA GAA AAA TGC TGA ATGA-3′; Reverse: 5′-CAG GAAACAGCTATGACACCACAATTGGGTGGA CA-3′). Every PCR reaction contained 12.5 μl 2 × Jump-Start REDTaq Ready Mix, 2 μl 10 μM primers (1 μl for each), 2 μl cDNA, and water to make a final volume of 25 μl. Cycling conditions were used as follows: 96 °C for 5 min, 94 °C for 30 s (35 ×), 58 °C for 45 s (35 ×), and 72 °C for 60 s (35 ×) with a final extension at 72 °C for 5 min (1 ×).
Fragment analysis
RT-PCR were performed using primers with the same sequences and PCR conditions as mentioned above in the cDNA analysis, except that the reverse primer was labeled with 5′JOE fluorophore. The RT-PCR products were subjected to fragment analysis on 3730 Genetic Analyzer (Applied Biosystems, Foster City, CA) using the internal lane standard 600 (ILS 600) (Promega Corporation, Madison, WI) as a DNA ladder to assign correct sizes to DNA fragments. The proportions of different transcripts were calculated based on the peak heights of each fragment.
Loss of heterozygosity (LOH) analysis in tumor tissue
Loss of heterozygosity analysis was performed in breast cancer tissue samples from the patient. Sections of the paraffin-embedded breast carcinoma sample were stained by hematoxylin and eosin (H&E) using standard protocols. Tumor cells were collected and deparaffinized. DNA extraction was performed using the Qiagen DNeasy Tissue Kit (Cat# 69,504, Qiagen). Tumor DNA was amplified using PCR (Forward primer: 5′-CTT CTC TCC ATT CCC CTG TC-3′; Reverse primer: 5′-CAT CGT GGG ATC TTG CTT AT-3′). PCR conditions consisted of initial denaturing at 96 °C for 5 min; 35 cycles of 94 °C for 30 s, 58 °C for 45 s, and 72 °C for 60 s; and final extension at 72 °C for 5 min. The PCR products were run on QIAxcel (Qiagen, Valencia, CA) to confirm successful amplification. The PCR products were subjected to direct DNA sequence analysis and fragment analysis performed by a 3730 DNA Analyzer (Applied Biosystems, Foster City, CA).
Results
The BRCA1 c. 5332+4delA variant disrupts normal mRNA splicing
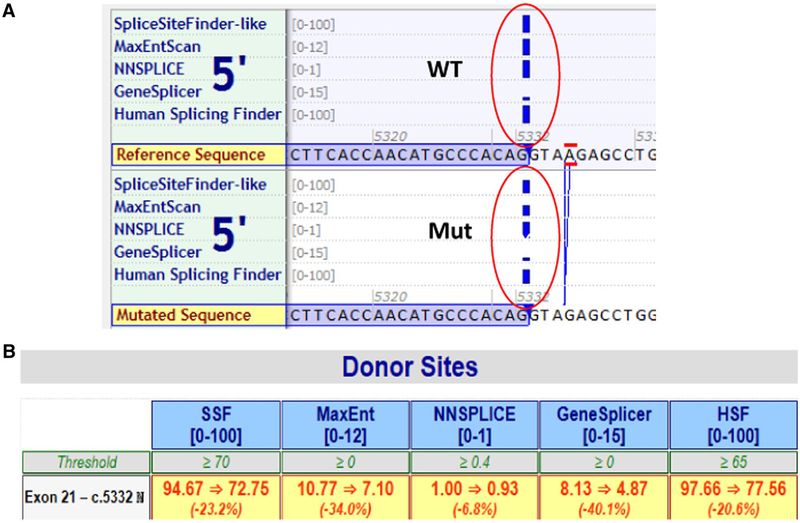
To evaluate the potential effects of the variant on splicing, we used Alamut software, which incorporates five tools to predict the potential effects of BRCA1 c.5332+4delA on normal mRNA splicing. Four out of five tools predicted that the variant significantly weakens the 5′ donor splice site by more than 20% (Fig. 2a, b).
Fig. 2.
In silico predictions of the BRCA1 c.5332+4delA variant. The Alamut software was used to evaluate the potential effects of the variant on splicing. Four out of five tools predicted that the variant weakens the 5′ donor splice site, causing an average 25% loss of function with estimates ranging from a loss of 7–40%
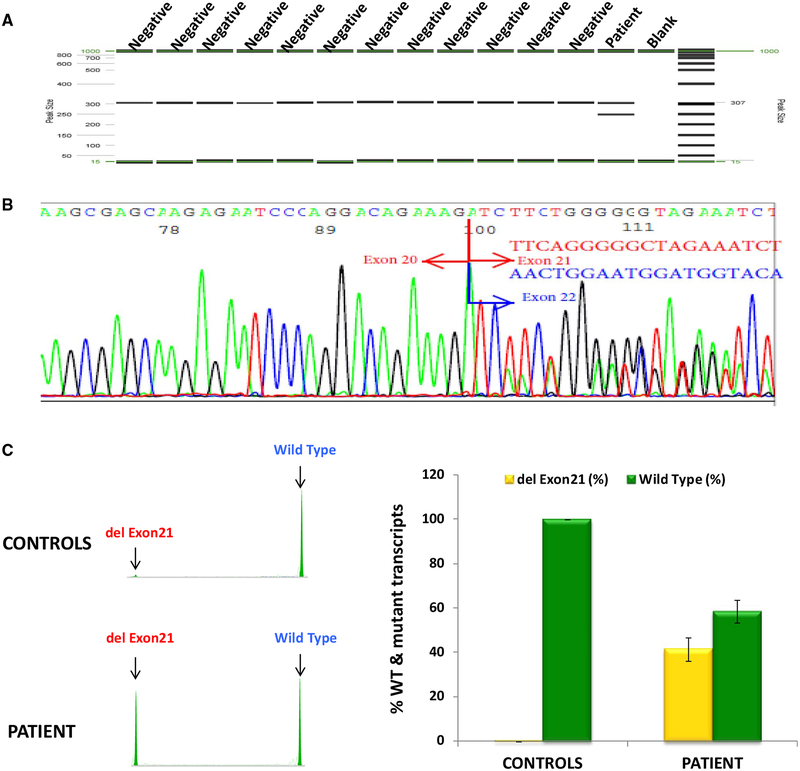
The effect of BRCA1 c. 5332+4delA variant on RNA splicing was subsequently evaluated by amplifying regions of BRCA1 from cDNA derived from the patient. PCR was designed to generate a fragment that spanned part of exon 19, the entire coding region of exons 20–22, and part of exon 23, which are likely affected by the variant. An additional band, which is absent in 12 controls, was identified in the patient (lane 14, Fig. 3a). This band represents an aberrant RNA splicing product attributable to this variant. Further sequencing revealed that this variant leads to loss of the entire exon 21, which contains 55 nucleotides (Fig. 3b). Semi-quantitative analysis from 5 patient replicates and 12 negative controls indicated that this aberrant RT-PCR product accounts for about 40% of the total transcript levels (Fig. 3c). We also performed BRCA1 full gene sequencing on patient sample to identify potential SNPs that might be used to address whether the mutant allele completely disrupts normal splicing. Unfortunately, we did not find any SNPs in the exonic regions and were unable to perform SNP tagging to exclude the possibility that the mutant allele still produces full-length transcript.
Fig. 3.
RT-PCR analysis demonstrates BRCA1 c.5332+4delA leads to exon 21 skipping. a RT-PCR products run on QIAxcel Advanced System from QIAGEN. An extra band was observed in the patient, but not in controls. b Electropherogram from ABI 3730xl DNA analyzer showing that the variant causes exon 21 skipping. The boundary of exons is marked by red arrow. c Semi-quantitative fragment analysis of RNA transcripts from the patient and controls
Loss of heterozygosity (LOH) analysis in tumor tissue
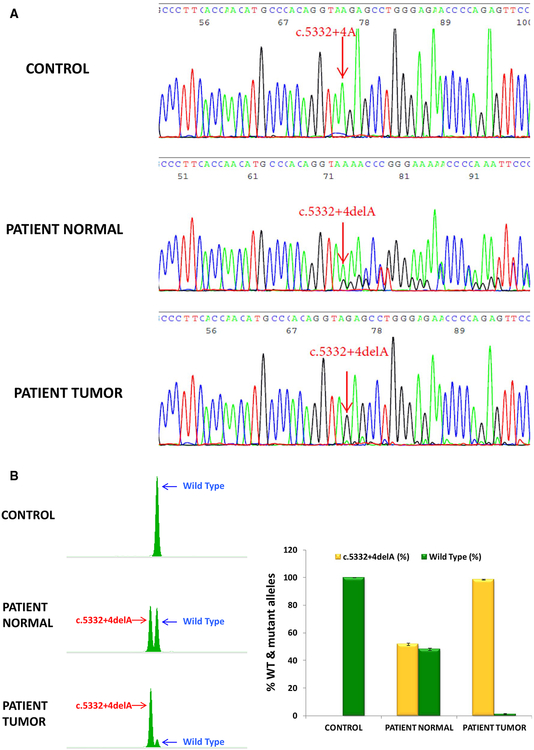
To investigate whether the patient’s tumor sample exhibits loss of the wild-type allele, DNA was extracted from the patient’s tumor and amplified along with DNA from both the patient’s normal tissue (peripheral blood) and a normal control sample that does not carry the BRCA1 variant. PCR fragments were sequenced (Fig. 4a) and analyzed using fragment analysis (Fig. 4b). As expected, patient’s normal tissue showed two alleles of approximately equal peak heights indicating heterozygous status (Fig. 4a, b). In the tumor sample, the c.5332+4delA allele was the major allele, and the peak height of the wild-type c.5332+4A allele was significantly reduced compared with that in patient’s normal samples (Fig. 4a, b). Fragment analysis was used to quantify the percentage of LOH (Fig. 4b). The patient DNA from normal blood comprises of both the wild-type and mutant alleles accounting for approximately 48.15 and 51.85% of the total transcript levels. In contrast, in the tumor sample, the wild-type allele significantly decreased to 1.5% and the c.5332+4delA mutant increased to 98.5%. LOH is defined as a signal reduction of 25% or more of one allele in the tumor sample, when compared with the normal sample, and was calculated as follows: [100% − percentage of wild-type allele in tumor tissue/percentage of wild-type allele in normal tissue] = [100% − 1.5%/48.15% = 97%]. The percentage of the wild-type allele in the above equation was calculated as an average of peak height × peak area of wild-type allele/sum of peak heights × peak areas of wild-type and mutant alleles from at least six independent experiments. Our data showed 97% signal reduction of the wild-type allele in the tumor compared to control samples demonstrating the loss of wild-type BRCA1 allele in the tumor.
Fig. 4.
Loss of heterozygosity (LOH) analysis demonstrates the loss of the wild-type allele in patient’s tumor tissue. a Sequencing analysis showing the peak height of the wild-type c.5332+4A allele was significantly reduced in the patient tumor compared with that in patient’s normal samples. b Quantification of the percentage of the wild-type and mutant alleles in normal and tumor tissue by fragment analysis. Left panel: peaks of the wild-type and mutant alleles analyzed by GeneMapper. Right panel: The percentage of the wild-type or mutant allele in control, patient normal tissue (blood), and patient tumor tissue
Discussion
This variant, BRCA1c.5332+4delA, affecting the donor splice site at the exon–intron 21 junction was identified in an individual with early-onset ER+/PR+/HER2− breast cancer with limited family history. The external commercial laboratory classified it as a VUS which was deposited into ClinVar. This rare variant is absent from the population databases and our cohort of more than 15,000 individuals undergoing germline testing. However, this variant was identified in 1/478 individuals with early-onset, high-risk breast cancer [26]. Further studies demonstrated this variant segregates with breast and ovarian cancer in the family. Experimental studies showed exon skipping similar to our observations, and a functionally impaired protein [27]. BRCA1 mutation carriers are typically diagnosed with high grade, early-onset, triple-negative breast cancers (TNBC) [28, 29] and morphological features can be useful in interpreting variants [30]; however, cases of early-onset breast cancer that are not TNBC have been reported. Interestingly, the splice site variant c.5332+1G>A, which affects the same donor site as c.5332+4delA, was observed de novo in an individual with bilateral early-onset breast cancer with a pathological profile similar to our case (ER+/PR+) [20]. Functional studies using patient-derived DNA showed c.5332+1G>A which resulted in full-length and exon 21-skipped RT-PCR products [19], similar to c.5332+4delA, and was classified pathogenic by ENIGMA. It is worth mentioning neither parent of our proband carries the BRCA1 c.5332+4delA variant, which indicates it is likely a de novo mutation (paternity testing not performed).
Alternatively spliced isoforms of BRCA1 have been identified in normal tissues, particularly isoforms skipping all or part of exon 11 [16, 17, 31]. The relative levels of these alternatively spliced isoforms appear to influence DNA synthesis and cancer cell proliferation. At least one of these isoforms appeared to regulate cell proliferation by altering subcellular localization. Naturally occurring isoforms missing exon 21 are uncommon; we detected less than 1% of alternatively spliced transcripts with exon 21 deleted in our control samples. Similarly, minimal levels of alternatively spliced transcripts with exon 21 deletion were seen in the control sample for the pathogenic c.5332+1G>A variant [19] although this was not quantified. The deletion of exon 21 would cause a frameshift that presumably creates a premature stop codon and leads to the loss of 90 amino acids within BRCA1 BRCT domain. We observed LOH of the variant allele in our patient’s tumor sample, which would be expected for a pathogenic variant. While it is commonly observed in breast cancers, LOH is not currently included in the criteria for variant classification according to ACMG guidelines [32].
This work highlights the lack of concordance sometimes seen between computational algorithms and experimental studies on analyzing splice variants, and the implications of in silico prediction in interpretation of intronic and synonymous variants. While five algorithms predict c.5332+4delA causes loss of function at the donor splice site 4 bp upstream, the predicted loss averages 25%, with the mutated sequence predicted to be significantly stronger than any cryptic donor sites in the vicinity. The functional evidence from our study suggests the effect is much more pronounced with a 60:40 ratio of full length: exon 21-skipped transcripts in our patient sample, similar to the results from c.5332+1G>A which is predicted to cause 100% loss of function at the canonical donor site [19]. Our observations are in line with the potential effects caused by the disruption of both the + 4 and + 5 positions of a consensus 5′ splice site sequence, which are predominantly AG [33, 34]. Several other BRCA1 variants affecting these sites have been reported pathogenic by multiple labs in the ClinVar database, including variants at c.4986+4A/+5G, c.5152+4A/+5G, c.5406+4A/+5G, and c.8754+4A/+5G, all of which alter the A or G nucleotides at the + 4 or + 5 sites, respectively. One limitation of this study is the absence of other SNVs in the entire BRCA1 coding region observed by BRCA1 full gene sequencing in our patient, precluding SNP tagging analysis to determine whether the mutant allele can generate normal transcripts.
Advances in NGS technology have facilitated the generation of large amounts of potentially actionable genomic data, but have also driven home the inherent difficulty in interpreting novel variants. This is particularly true for novel intronic variants beyond the ± 1~2 canonical splice sites as well as synonymous variants, which do not typically impact protein structure but may affect gene function through disruption of normal splicing. Functional studies are necessary to determine pathogenicity of these variants.
Conclusions
We identified and characterized a splice site variant, c.5332+4delA, in an individual with early-onset breast cancer that affects splicing resulting in the loss of exon 21—an isoform that is not a naturally occurring alternatively spliced BRCA1 isoform, supporting a Likely Pathogenic classification according to ACMG guidelines [32].
Funding
Funds for this study were provided by the Department of Pathology, Memorial Sloan Kettering Cancer Center.
References
- 1.Zhang J, Powell SN (2005) The role of the BRCA1 tumor suppressor in DNA double-strand break repair. Mol Cancer Res 3(10):531–539 [DOI] [PubMed] [Google Scholar]
- 2.Scully R, Ganesan S, Vlasakova K, Chen J, Socolovsky M, Livingston DM (1999) Genetic analysis of BRCA1 function in a defined tumor cell line. Mol Cell 4(6):1093–1099 [DOI] [PubMed] [Google Scholar]
- 3.Koonin EV, Altschul SF, Bork P (1996) BRCA1 protein products functional motifs. Nat Genet 13(3):266–268 [DOI] [PubMed] [Google Scholar]
- 4.Williams RS, Green R, Glover JN (2001) Crystal structure of the BRCT repeat region from the breast cancer-associated protein BRCA1. Nat Struct Biol 8(10):838–842 [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez JA, Au WW, Henderson BR (2004) Cytoplasmic mis-localization of BRCA1 caused by cancer-associated mutations in the BRCT domain. Exp Cell Res 293(1):14–21 [DOI] [PubMed] [Google Scholar]
- 6.Friedman LS, Ostermeyer EA, Szabo CI, Dowd P, Lynch ED, Rowell SE, King MC (1994) Confirmation of BRCA1 by analysis of germline mutations linked to breast and ovarian cancer in ten families. Nat Genet 8(4):399–404 [DOI] [PubMed] [Google Scholar]
- 7.Futreal PA, Liu Q, Shattuck-Eidens D, Cochran C, Harshman K, Tavtigian S, Bennett LM, Haugen-Strano A, Swensen J, Miki Y et al. (1994) BRCA1 mutations in primary breast and ovarian carcinomas. Science 266(5182):120–122 [DOI] [PubMed] [Google Scholar]
- 8.Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A et al. (2003) Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet 72(5):1117–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King MC, Marks JH, Mandell JB (2003) New York Breast Cancer Study G: breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science 302(5645):643–646 [DOI] [PubMed] [Google Scholar]
- 10.van der Kolk DM, de Bock GH, Leegte BK, Schaapveld M, Mourits MJ, de Vries J, van der Hout AH, Oosterwijk JC (2010) Penetrance of breast cancer, ovarian cancer and contralateral breast cancer in BRCA1 and BRCA2 families: high cancer incidence at older age. Breast Cancer Res Treat 124(3):643–651 [DOI] [PubMed] [Google Scholar]
- 11.Mavaddat N, Peock S, Frost D, Ellis S, Platte R, Fineberg E, Evans DG, Izatt L, Eeles RA, Adlard J et al. (2013) Cancer risks for BRCA1 and BRCA2 mutation carriers: results from prospective analysis of EMBRACE. J Natl Cancer Inst 105(11):812–822 [DOI] [PubMed] [Google Scholar]
- 12.Larsen MJ, Thomassen M, Gerdes AM, Kruse TA (2014) Hereditary breast cancer: clinical, pathological and molecular characteristics. Breast Cancer 8:145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheon JY, Mozersky J, Cook-Deegan R (2014) Variants of uncertain significance in BRCA: a harbinger of ethical and policy issues to come? Genome Med 6(12):121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baralle D, Baralle M (2005) Splicing in action: assessing disease causing sequence changes. J Med Genet 42(10):737–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perrin-Vidoz L, Sinilnikova OM, Stoppa-Lyonnet D, Lenoir GM, Mazoyer S (2002) The nonsense-mediated mRNA decay pathway triggers degradation of most BRCA1 mRNAs bearing premature termination codons. Hum Mol Genet 11(23):2805–2814 [DOI] [PubMed] [Google Scholar]
- 16.Colombo M, Blok MJ, Whiley P, Santamariña M, Gutiérrez-Enríquez S, Romero A, Garre P, Becker A, Smith LD, De Vecchi G et al. (2014) Comprehensive annotation of splice junctions supports pervasive alternative splicing at the BRCA1 locus: a report from the ENIGMA consortium. Hum Mol Genet 23(14):3666–3680 [DOI] [PubMed] [Google Scholar]
- 17.Orban TI, Olah E (2003) Emerging roles of BRCA1 alternative splicing. Mol Pathol 56(4):191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spurdle AB, Healey S, Devereau A, Hogervorst FBL, Monteiro ANA, Nathanson KL, Radice P, Stoppa-Lyonnet D, Tavtigian S, Wappenschmidt B et al. (2012) ENIGMA—Evidence-based network for the interpretation of germline mutant alleles: an international initiative to evaluate risk and clinical significance associated with sequence variation in BRCA1 and BRCA2 genes. Hum Mutat 33(1):2–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colombo M, De Vecchi G, Caleca L, Foglia C, Ripamonti CB, Ficarazzi F, Barile M, Varesco L, Peissel B, Manoukian S et al. (2013) Comparative in vitro and in silico analyses of variants in splicing regions of BRCA1 and BRCA2 genes and characterization of novel pathogenic mutations. PLoS ONE 8(2):e57173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edwards E, Yearwood C, Sillibourne J, Baralle D, Eccles D (2009) Identification of a de novo BRCA1 mutation in a woman with early onset bilateral breast cancer. Fam Cancer 8(4):479–482 [DOI] [PubMed] [Google Scholar]
- 21.Steffensen AY, Dandanell M, Jonson L, Ejlertsen B, Gerdes AM, Nielsen FC, Hansen T (2014) Functional characterization of BRCA1 gene variants by mini-gene splicing assay. Eur J Hum Genet 22(12):1362–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Casado Z, Romero I, Fernandez-Serra A, Rubio L, Llopis F, Garcia A, Llombart P, Lopez-Guerrero JA (2011) A de novo complete BRCA1 gene deletion identified in a Spanish woman with early bilateral breast cancer. BMC Med Genet 12:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen W, Pan K, Ouyang T, Li J, Wang T, Fan Z, Fan T, Lin B, Lu Y, You W et al. (2009) BRCA1 germline mutations and tumor characteristics in Chinese women with familial or early-onset breast cancer. Breast Cancer Res Treat 117(1):55–60 [DOI] [PubMed] [Google Scholar]
- 24.Li WF, Hu Z, Rao NY, Song CG, Zhang B, Cao MZ, Su FX, Wang YS, He PQ, Di GH et al. (2008) The prevalence of BRCA1 and BRCA2 germline mutations in high-risk breast cancer patients of Chinese Han nationality: two recurrent mutations were identified. Breast Cancer Res Treat 110(1):99–109 [DOI] [PubMed] [Google Scholar]
- 25.Ahlborn LB, Dandanell M, Steffensen AY, Jonson L, Nielsen FC, Hansen TV (2015) Splicing analysis of 14 BRCA1 missense variants classifies nine variants as pathogenic. Breast Cancer Res Treat 150(2):289–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park B, Sohn JY, Yoon K-A, Lee KS, Cho EH, Lim MC, Yang MJ, Park SJ, Lee MH (2017) Characteristics of BRCA1/2 mutations carriers including large genomic rearrangements in high risk breast cancer patients. Breast Cancer Res Treat 163(1):139–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon K-A, Kong S-Y, Lee EJ, Cho JN, Chang S, Lee ES (2017) A novel germline mutation in BRCA1 causes exon 20 skipping in a Korean family with a history of breast cancer. J Breast Cancer 20(3):310–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peshkin BN, Alabek ML, Isaacs C (2010) BRCA1/2 mutations and triple negative breast cancers. Breast Dis. 10.3233/BD-2010-0306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuchenbaecker KB, Ramus SJ, Tyrer J, Lee A, Shen HC, Beesley J (2015) Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat Genet 47:164–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hofstra RMW, Spurdle AB, Eccles D, Foulkes WD, de Wind N, Hoogerbrugge N, Hogervorst FBL (2008) Tumor characteristics as an analytic tool for classifying genetic variants of uncertain clinical significance. Hum Mutat 29(11):1292–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tammaro C, Raponi M, Wilson David I, Baralle D (2012) BRCA1 exon 11 alternative splicing, multiple functions and the association with cancer. Biochem Soc Trans 40(4):768–772 [DOI] [PubMed] [Google Scholar]
- 32.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E et al. (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17(5):405–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roca X, Krainer AR, Eperon IC (2013) Pick one, but be quick: 5′ splice sites and the problems of too many choices. Genes Dev 27(2):129–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sibley CR, Blazquez L, Ule J (2016) Lessons from non-canonical splicing. Nat Rev Genet 17(7):407–421 [DOI] [PMC free article] [PubMed] [Google Scholar]