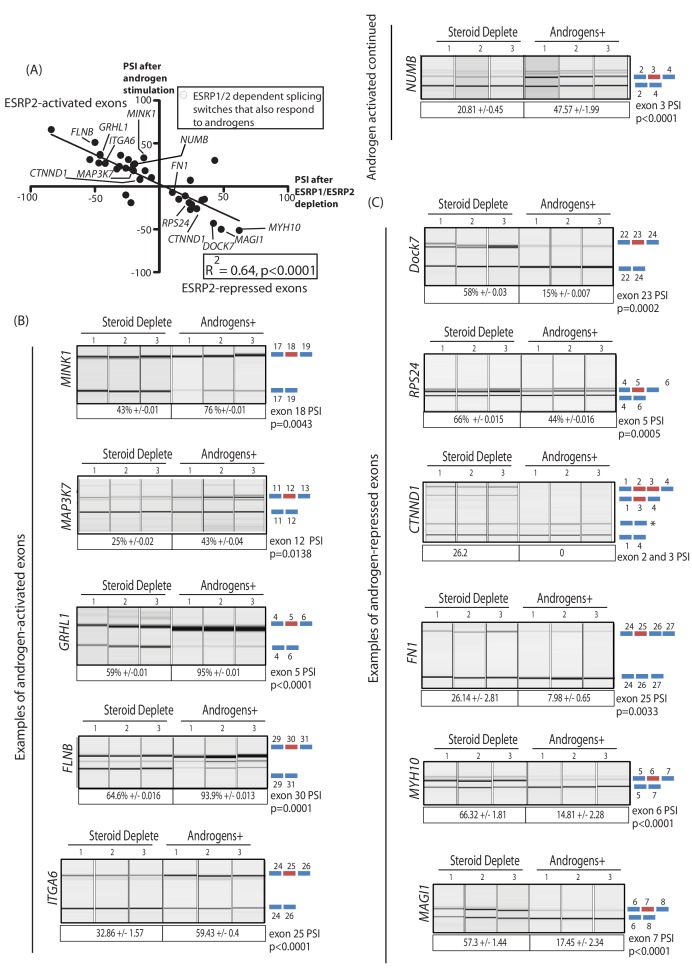
Figure 4. An androgen steroid hormone-ESRP2 axis controls alternative splicing in prostate cancer cells.
(A) ESRP2-regulated exons are frequently also controlled by androgens in prostate cancer cells. 31/48 of the ESRP target exons (identified by RNAseq analysis of LNCaP cells depleted of ESRP1 and ESRP2) were regulated in the opposite direction in LNCaP cells treated by androgens (10nM R1881) for 48 hr (which would induce ESRP2 protein expression). Plotting the splicing responses to androgen stimulation with those after ESRP1/ESRP2 depletion revealed a negative correlation (slope = −0.66+/−0.09, Rsquare = 0.64, p<0.0001, calculated using Graphpad). Individual values for this graph are given in Figure 3—source data 2, and are averages from three biological replicates. (B) Capillary gel electrophoretograms showing splicing patterns of 3 biological replicates grown in steroid deplete media, or steroid deplete media supplemented with R1881, for alternative exons that are activated by androgen exposure in the MINK1, MAP3K1, GRHL1, FLNB, ITGA6 and NUMB genes. (C) Capillary gel electrophoretograms showing splicing patterns of 3 biological replicates grown in steroid deplete media, or steroid deplete media supplemented with R1881, for alternative exons that are repressed by 48 hr androgen exposure in the DOCK7, RPS24, CTNND1, FN1, MYH10 and MAGI1 genes that were repressed by four androgen treatment. For parts (B–C) the p values were calculated using unpaired t tests, apart for CTNND1 where zero inclusion of exons 2 and 3 were detected in the presence of androgens. For CTNND1, an RT-PCR product derived from a splice variant joining exons 1–4 via an alternative splice site is asterisked.