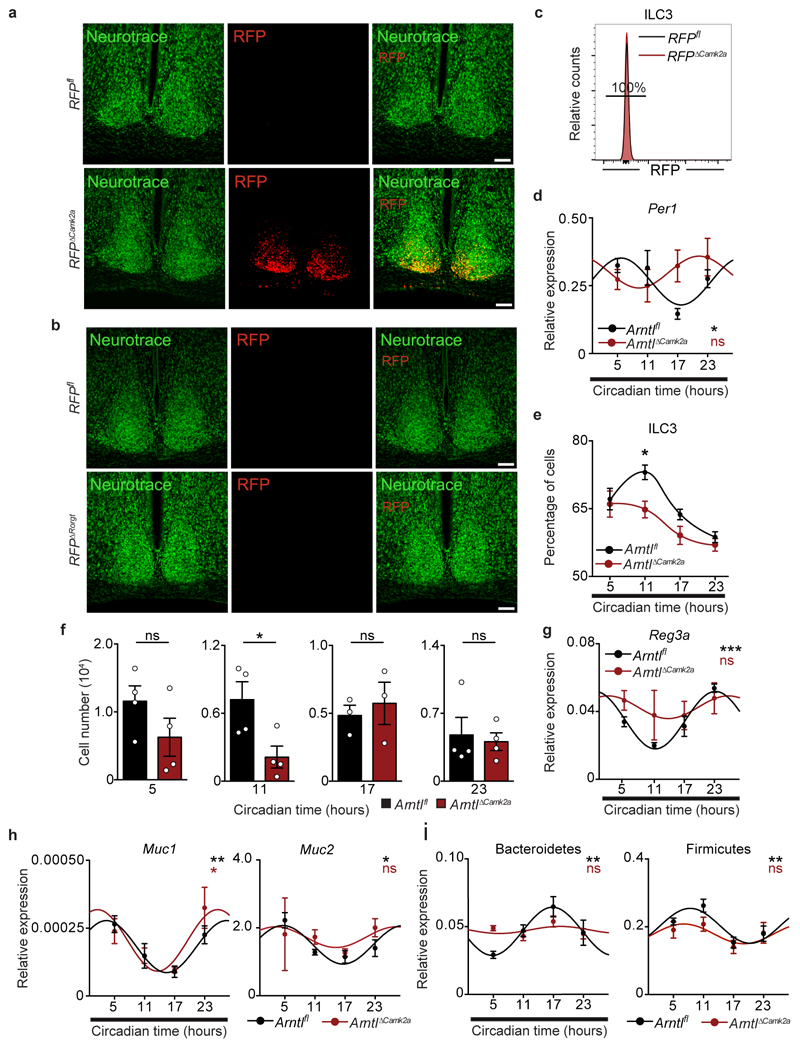
Extended Data Figure 9. Brain-tuned signals shape gut ILC3.
a, b, Confocal image of coronal brain sections showing neurotrace and RFP expression in the SCN. Scale bar: 100 µm. Representative of 3 independent analyses. c, Representative histogram of RFP expression in small intestine lamina propria ILC3. Representative of 3 independent analyses. d, Per1 expression in small intestinal lamina propria ILC3. n=3. e, Percentage of small intestine lamina propria ILC3. n=3. f, Number of small intestine lamina propria ILC3. n=3. g, h, Epithelial reactivity gene expression in the intestinal epithelium. n=3. i, Rhythms of faecal Bacteroidetes and Firmicutes. Arntlfl n=4, ArntlΔCamk2a n=3. (a-i) n represents biologically independent animals. Mean and error bars: s.e.m.. (d,g-i) Cosinor regression was used to define circadian rhythmicity; Cosine fitted curves are shown. (e) two-way ANOVA and Sidak’s test; (f) two-tailed unpaired Student t-test; *P<0.05; **P<0.0;1; ns not significant.