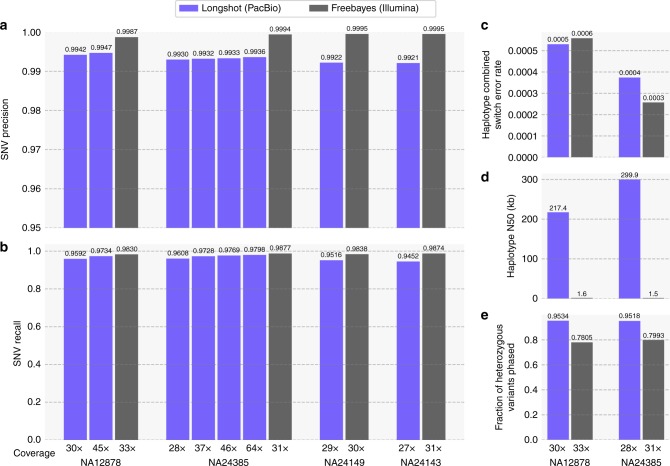
Fig. 2.
Accuracy and completeness of LongShot SNV calls on whole-genome SMS data. Longshot was used to call single nucleotide variants (SNVs) using SMS data from the GIAB project for four human genomes: NA12878 ( and coverage), NA24385 (, , , and coverage), NA24149 ( coverage), and NA24143 ( coverage). For each individual, variants were also called using FreeBayes applied to coverage Illumina short reads. a Precision of the SNV calls calculated using the GIAB high-confidence variant call set. b Recall of the SNV calls. c The combined switch error rate (total rate of switch errors and mismatch errors) of the Longshot and Illumina short-read-based haplotypes. d N50 length of the haplotypes. e The fraction of heterozygous variants phased in each dataset