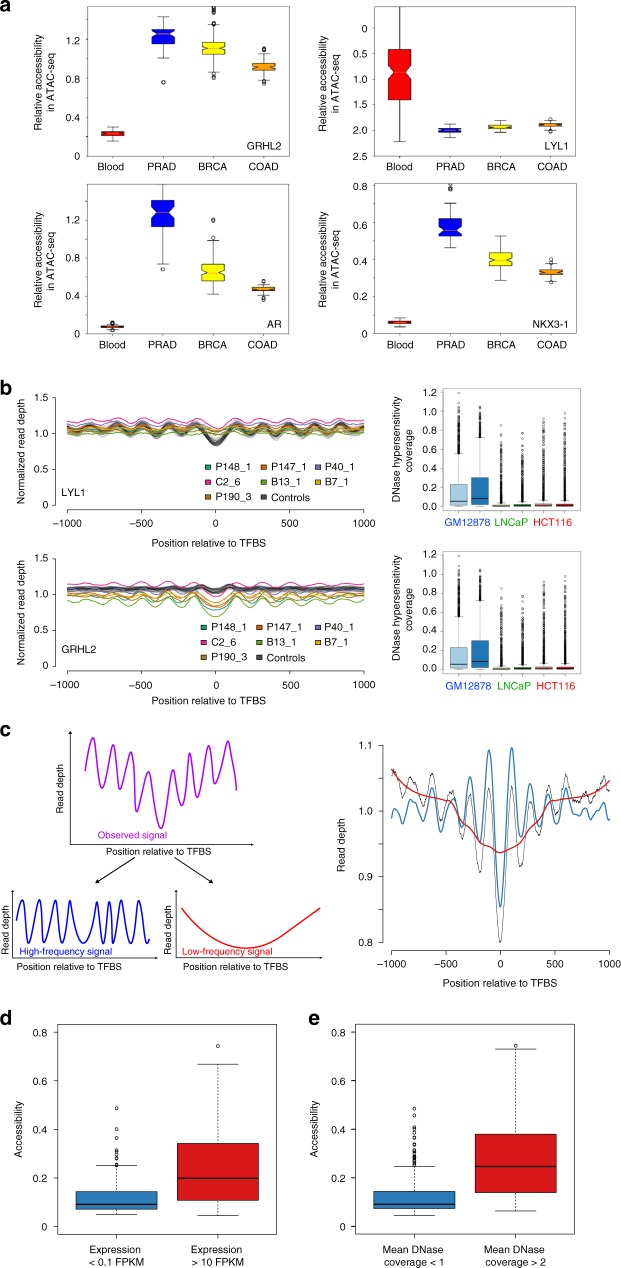
Fig. 1.
Bioinformatic fragment analysis of cfDNA enables accurate inference of TF activity. a Relative accessibility for TFs GRHL2, LYL1, AR, and NKX3-1 for hematopoietic, PRAD, BRCA, and COAD lineages generated from publicly available ATAC-seq data12,13. b Nucleosome position profiles from plasma DNA for the hematopoietic lineage TF LYL1 and the epithelial TF GRHL2 (left panels; profiles of healthy controls are shown in gray, and patient-derived profiles are displayed in the indicated colors). In plasma samples from patients with cancer, the amplitudes of LYL1 and GRHL2 are decreased and increased, respectively, reflecting the different contributions of DNA from hematopoietic and epithelial cells to the blood. The lineage specificity was further confirmed by DNA hypersensitivity assays (right panels). c To measure TF accessibility, the observed raw coverage signal (purple in left and black in right panel) was split by Savitzky–Golay filtering into a high-frequency signal (blue) and a low-frequency signal (red) using different window sizes. The right panel illustrates an overlay of these three signals. The high-frequency signal is used as a measure for accessibility. d TFs with increased expression (n = 137; >10 FPKM [fragments per kilobase exon per million reads]) in blood have a significantly higher accessibility as compared with TFs with no or low signs of expression (n = 91; <0.1 FPKM). e Transcription factors with a mean DNase hypersensitivity coverage of >2 in GM12878 DNase data from the ENCODE project (n = 213) have higher accessibilities than factors that have a mean coverage of <1 (n = 244). Boxplots are defined as follows: Centerline represents the median, the box represents the interquartile range (IQR), and the whiskers denote the first quartile –1.5 IQR and the third quartile +1.5 IQR, respectively. Notches in (a) represent the confidence interval around the median