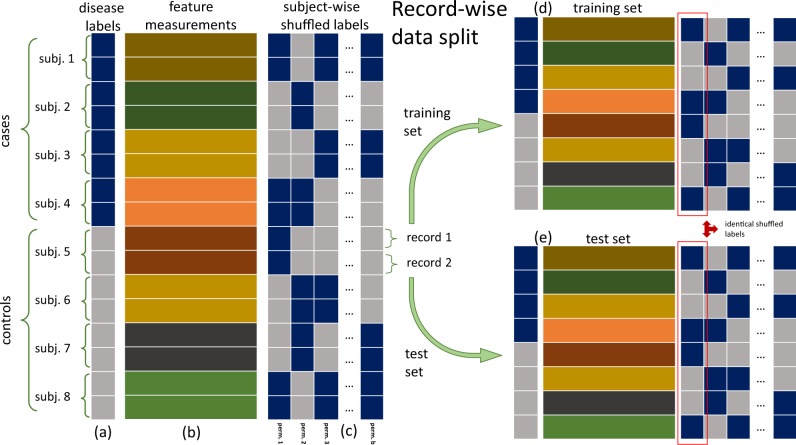
Fig. 1.
Permutation scheme to detect identity confounding. The schematic shows a toy example for a data-set with eight subjects (four cases and four controls), where each subject contributed two records. a and b show, respectively, the disease label vector and the feature matrix. c Shows distinct “subject-wise random permutations” of the disease labels, where the permutations are performed at the subject level, rather than at the record level (so that all records of a given subject are assigned either “case” or “control” labels). For example, in the first permutation, the labels of subjects 2 and 3 changed from “case” to “control”, the labels of subjects 5 and 8 changed from “control” to “case”, and the labels of subjects 1, 4, 6, and 7 remained the same. (Note that for each subject, the labels are changed across all records). The subject-wise label permutations destroy the association between the disease labels and the features, making it impossible for a classifier trained with shuffled labels to learn the disease signal. Adopting the record-wise data split strategy, with half of the records assigned to the training set (d), and the other half to the test set (e), we have that both training and test sets contain 1 record from each subject. Most importantly, in each permutation the shuffled labels of each subject are the same in both the training and test sets. For instance, in the first permutation (highlighted by the red boxes in d, e) we have that the shuffled labels of subjects 1 to 8, namely, “case”, “control”, “control”, “case”, “case”, “control”, “control”, “case”, are exactly the same in the training and test sets. Consequently, any classifier trained with the shuffled labels will still be able to learn to identify individuals, even though it cannot learn the disease signal