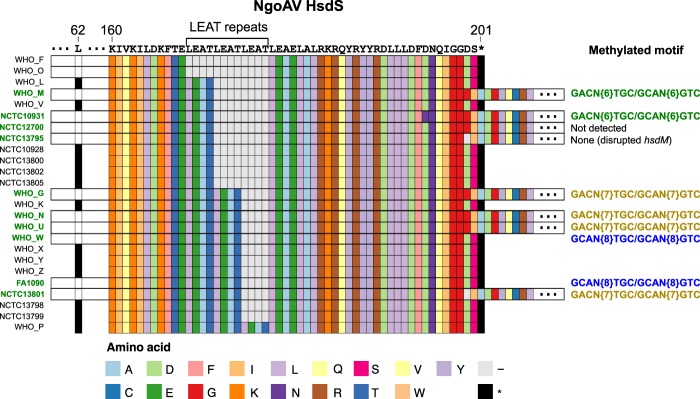
Figure 3.
Protein alignment of the HsdS specificity unit of the Type I NgoAV Restriction-Modification system (RMS). The three main regions of variation within the unit are shown as a premature stop codon in position 62 of the protein that causes inactivation of the RMS, a variable number of LEAT repeats and a downstream stop codon in position 201 (caused by a change in the length of a poly-G homopolymer) which, in combination, are related to a different recognition motif. The strains with a full-length protein are labelled in green (left), and the methylated motifs detected using PacBio are shown in different colours on the right. ‘−‘ represents gaps and ‘*’ stop codons.