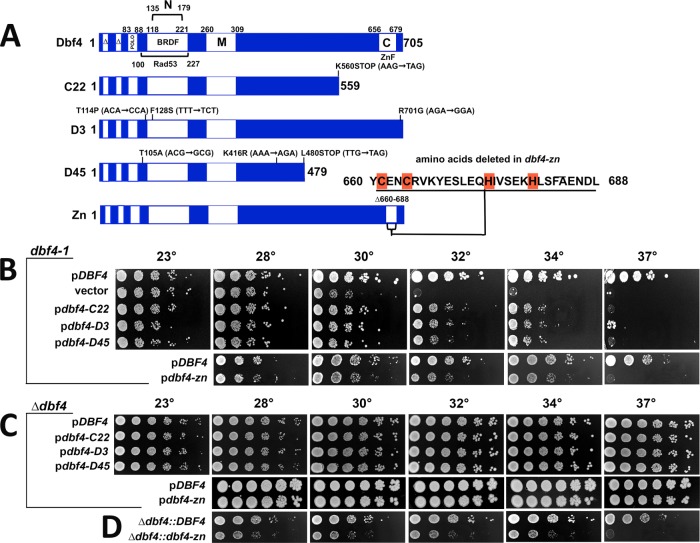
FIGURE 2:
Characterization of dbf4-C and dbf4-zn. Experiments in this figure are without HU treatment. (A) Dbf4 domains and mutant alleles. C22, D3, and D45 are PCR mutagenized alleles, while dbf4-zn was constructed using recombinant techniques. Base pair changes that alter amino acid coding in dbf4-C alleles are indicated; Zn2+ finger amino acids deleted in dbf4-zn are also shown. The diagram also illustrates amino acid boundaries for domains involved in cell cycle proteolysis (∆); Cdc5 (Polo) and Rad53 binding; motifs N, M, and C; and the BRCT-related BFDF domain. (B) Complementation of dbf4-1; 10-fold serial dilutions of a dbf4-1 strain (JBY997) transformed with low copy pCEN ARS DBF4 (JJY059; JJY164), vector (JJY016), pdbf4-C22 (JJY017), pdbf4-D3 (JJY060), pdbf4-D45 (JJY061), and pdbf4-zn (JJY165) plasmids were stamped onto selective media at indicated temperatures. Whereas DBF4 complements dbf4-1 to 37°C, dbf4-C and dbf4-zn alleles partially complement to 34°C. (C) Complementation of dbf4-∆. A dbf4-∆ strain harboring pURA3 DBF4 was transformed with pCEN ARS DBF4 (JJY037, JJY166), pdbf4-C22 (JJY032), pdbf4-D3 (JJY033), pdbf4-D45 (JJY044), and pdbf4-zn (JJY167). The transformants were cured of the covering DBF4 URA3 plasmid on 5′-FOA; 10-fold serial dilutions were stamped onto selective media at indicated temperatures. In this case, low copy dbf4-C and dbf4-zn plasmids complement dbf4-∆ growth to 37°C. (D) Integrated dbf4-zn. Constructs expressing DBF4 (JJY046) or dbf4-zn (JJY076) were integrated immediately upstream of a precise deletion of the DBF4 open reading frame; 10-fold serial dilutions were incubated at indicated temperatures revealing a dbf4-zn temperature sensitive growth phenotype.