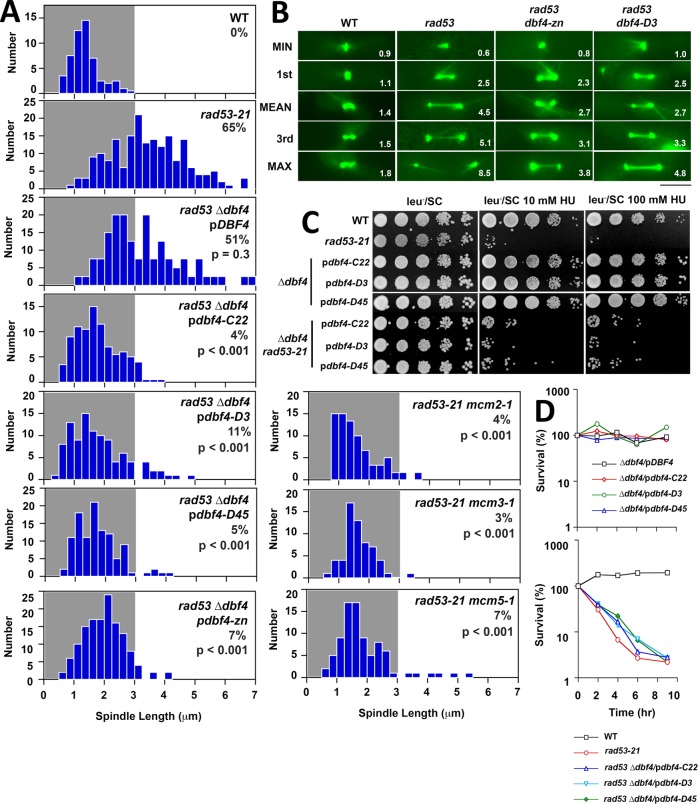
FIGURE 4:
pdbf4 C, pdbf4-zn and mcm alleles suppress rad53 spindle extension in HU. (A) WT (JBY1129), rad53-21 (JBY1274), rad53-21 dbf4-∆/pDBF4 (JJY184), rad53-21 dbf4-∆/pdbf4-C22 (JJY028), rad53-21 dbf4-∆/pdbf4-D3 (JJY029), rad53-21 dbf4-∆/pdbf4-D45 (JJY030), rad53-21 dbf4-∆/pdbf4-zn (JJY182), rad53-21 mcm2-1 (JJY112), rad53-21 mcm3-1 (JJY117), and rad53-21 mcm5-1 (JJY120) strains, all harboring SPC42-GFP, were released from G1 into 200 mM HU at 30°C. After 90 min spindle lengths were measured in fixed cells. Percentages of cells with spindles ≥3 μm are displayed, along with the results of two-tailed t tests comparing each mutant to the rad53-21 distribution. (B) WT (JBY1129), rad53-21 (JBY1274), rad53-21 dbf4-∆/pdbf4-zn (rad53 dbf4-zn on the figure; JJY182), and rad53-21 dbf4-∆/pdbf4-D3 (rad53 dbf4-D3 on the figure; JJY029) strains were transformed with pGFP-TUB1 to visualize MTs. Cells were released from G1 into 200 mM HU at 30°C. Starting at 90 min, spindles were imaged and measured (values at the bottom right of each panel) in 50 live cells. Spindles corresponding to minimum, maximum, mean, first, and third quadrant measurements are shown. Bar, 4 μm. (C) Tenfold serial dilutions of WT (JBY1129), rad53-21 (JBY1274), dbf4-∆/pDBF4 (JJY037), dbf4-∆/pdbf4-C22 (JJY032), dbf4-∆/pdbf4-D3 (JJY033), and dbf4-∆/pdbf4-D45 (JJY044) strains, along with rad53-21 dbf4-∆ transformants described in A were stamped on indicated media at 30°C to evaluate HU sensitivity. (D) Asynchronous cultures of all the strains described in C were shifted into media containing 200 mM HU at 30°C. At the indicated times, plating efficiency was determined to evaluate HU recovery.