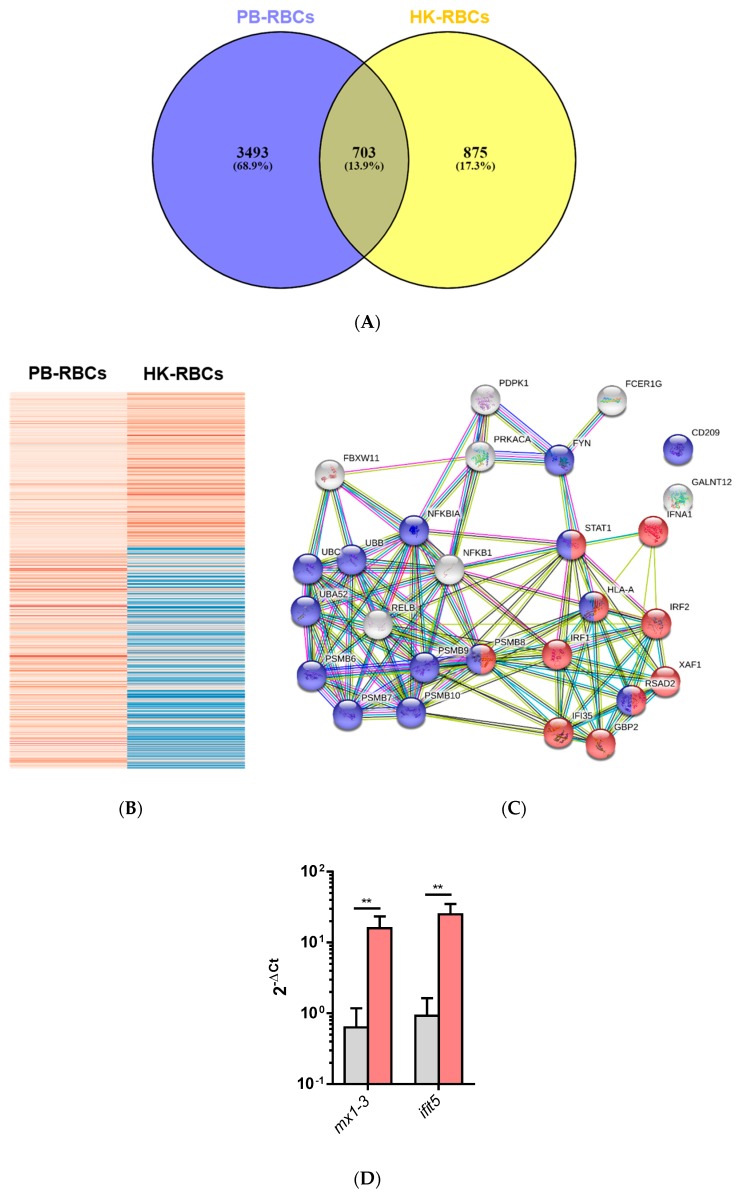
Figure 3.
Comparative overview of DEGs shared by HK-PBCs and PB-RBCs. (A) Venn diagram of DEGs in RBCs from each tissue. The middle region shows genes expressed in RBCs from both tissues. (B) Clustering of gene expression of common DEGs in HK-RBCs and PB-RBCs was performed using ClustVis. The dataset was inserted into matrix category. Parameters for clusterization included no data transformation and no row scaling, and the principal component analysis (PCA) method used was singular value decomposition (SVD) with imputation. Red indicates higher expression and blue represents lower expression. (C) PPI networks of the DEGs shared between PB-RBCs and HK-RBCs from VHSV-challenged rainbow trout identified with GO Immune System Process terms and constructed using STRING software (p-value < 10−16). Nodes represent proteins, while edges denote interaction between two proteins. Different line colors represent the types of evidence used in predicting the associations: Gene fusion (red), gene neighborhood (green), co-expression (black), gene co-occurrence (blue), experimentally determined (purple), from curated databases (teal), text-mining (yellow), or protein homology (lilac). Red nodes denote proteins implicated in the type I interferon signaling pathway (GO:0060337) and blue nodes denote proteins implicated in viral process (GO:0016032). (D) Expression of interferon-stimulated genes mx1-3 and ifit5 in PB-RBCs from VHSV-challenged rainbow trout (red bars) compared to mock-challenged (gray bars) as control at 2 dpc. Data represent mean ± SD (n = 6). A Mann–Whitney test was performed to test statistical significance between PB-RBCs from both groups. Asterisks denote statistical significance. ** p-value < 0.01.