Fig. 4.
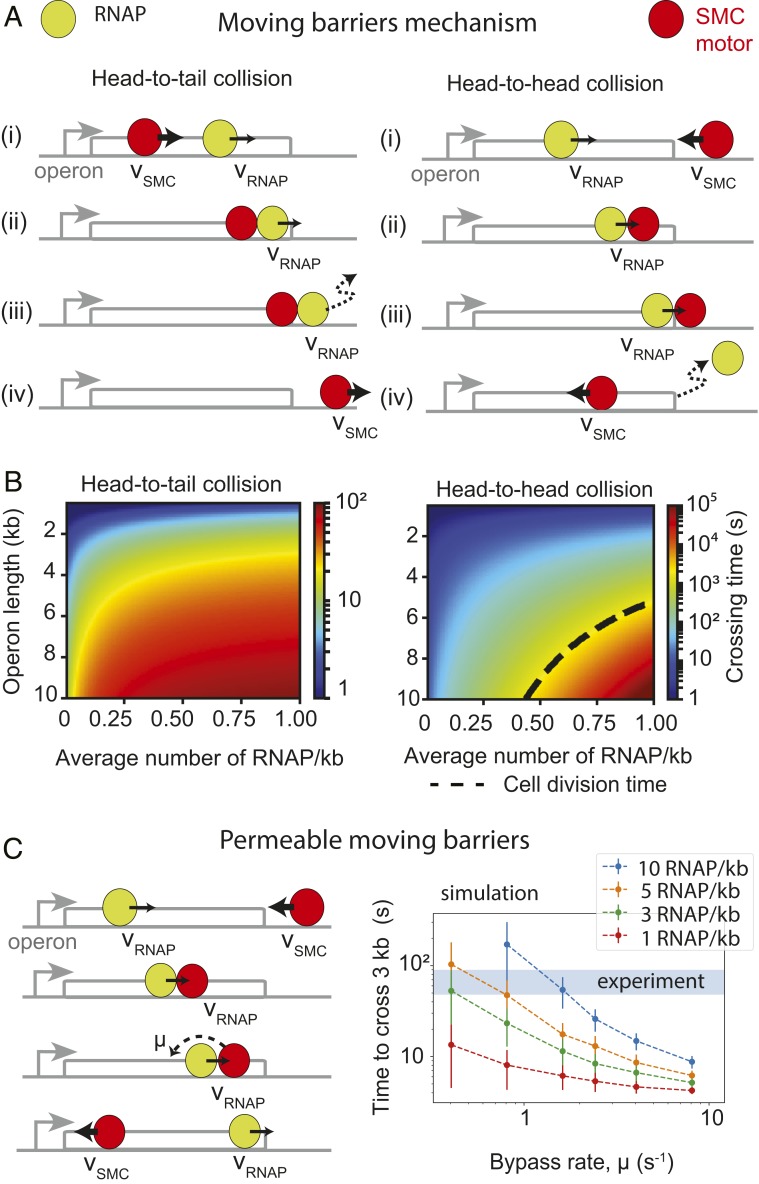
Mechanistic models of extrusion-transcription interference. (A) The “moving-barriers” model of condensin transcription interactions: Transcription complexes are posited as impermeable barriers to condensin movement, which results in different translocation dynamics when crossing operons in the co-oriented or convergent fashion. Condensin translocates (i) at its native speed (vSMC) until (ii) it reaches a slowly moving RNAP, then (iii) it proceeds in the direction and at the speed of RNAP (vRNAP) until (iv) RNAP reaches the end of the operon, whereby condensin continues translocation at its original speed and direction. (B) Analytically computed average times to cross an operon for each of the head-to-tail (Left) and head-to-head (Right) cases as a function of operon length and RNAP density. Despite similar “rules,” in the head-to-tail case (B, Left), the SMC always reaches the end of the operon, whereas in the head-to-head case (B, Right), a successful traversal by SMC may occur only if no RNAP is encountered within a cell-division time (dashed line). (C) Extension of the “moving-barriers” model allowing for condensin to bypass transcribing RNAP (Left, schematic); simulations of the locus-crossing times with varying permeability (“bypass”) rates and RNAP densities (Right); blue region indicates the experimentally estimated time for condensin to cross a 3-kb rRNA gene locus (with density ∼10 RNAP/kb).