Significance
Cross-species transmission of viruses may result in emergence of viruses that cause new pandemics. Most emerging infectious diseases are caused by RNA viruses with high mutation rates and plasticity of their genomes, which make them particularly fit to cross species barriers and adapt to new hosts. A DNA virus, canine parvovirus (CPV) provides a unique example of a pandemic virus that jumped species. Genetic and biochemical data revealed the genetic basis of the interaction between CPV and TfR; here, we use cryo-electron microscopy to provide icosahedral and asymmetric structures of the CPV–receptor complex, discovering a unique binding mode and a dynamic footprint of TfR on the capsid, solving the mystery behind the processes that allowed CPV to jump species.
Keywords: cryo-EM structure, CPV, TfR, dynamic, host jump
Abstract
Canine parvovirus (CPV) is an important pathogen causing severe diseases in dogs, including acute hemorrhagic enteritis, myocarditis, and cerebellar disease. Cross-species transmission of CPV occurs as a result of mutations on the viral capsid surface that alter the species-specific binding to the host receptor, transferrin receptor type-1 (TfR). The interaction between CPV and TfR has been extensively studied, and previous analyses have suggested that the CPV–TfR complex is asymmetric. To enhance the understanding of the underlying molecular mechanisms, we determined the CPV–TfR interaction using cryo-electron microscopy to solve the icosahedral (3.0-Å resolution) and asymmetric (5.0-Å resolution) complex structures. Structural analyses revealed conformational variations of the TfR molecules relative to the binding site, which translated into dynamic molecular interactions between CPV and TfR. The precise footprint of the receptor on the virus capsid was identified, along with the identity of the amino acid residues in the virus–receptor interface. Our “rock-and-roll” model provides an explanation for previous findings and gives insights into species jumping and the variation in host ranges associated with new pandemics in dogs.
Cross-species transmission plays a major role in virus evolution (1), and emerging viruses that overcame species barriers, including those due to variant cell receptors, can cause severe outbreaks in human populations; for example, severe acute respiratory syndrome, Nipah, and Ebola (2). Among the cross-species transmitting viruses, canine parvovirus (CPV) is a well-recognized example due to its pandemic spread, broad host range, and host-specific adaptation despite its DNA genome (3). CPV variants also infect numerous host animals besides dogs, mostly within the order Carnivora, including mink, raccoons, foxes, and black-backed jackals (bbjs). CPV type 2 (CPV-2), which originated from a virus closely related to feline panleukopenia virus (FPV), gained the host range for dogs in the mid-1970s and caused a CPV pandemic (4, 5). During 1979 and 1980, CPV-2 was completely replaced by a new variant, CPV-2a that is genetically and antigenically distinct from CPV-2 (6, 7). Currently, multiple CPV variants, including CPV-2a, are cocirculating worldwide. Extensive genetic and biochemical studies show that specific mutations displayed on or near the capsid surface alter the CPV binding phenotype to a host receptor, transferrin receptor type-1 (TfR), as well as the ability of the virus to infect many different hosts (6, 8, 9). Notably, many of the variations seen in the animal host ranges of CPV can be recapitulated by in vitro passage of the virus in different host cells (10).
CPV has a 26-nm diameter T = 1 icosahedral capsid that encapsidates a single-stranded DNA (ssDNA) genome of around 5,100 nucleotides. The capsid structures determined by X-ray crystallography show that the DNA-containing virions and empty capsids have nearly identical structures (11–13). The capsid shell comprises 60 copies of 2 viral proteins (VP1 and VP2), which are generated by differential mRNA splicing, with about 90% of the protein in the capsid being VP2 and 10% VP1. Each VP folds into an 8-stranded, antiparallel β-barrel structure, where the β-strands are connected by long loops that comprise most of the surface features of the capsid. At each 5-fold vertex of the icosahedral capsid surface, there is a cylinder with a 14- to 22-Å channel through which the N terminus of VP1 or the ssDNA genome may pass (11). Around the 5-fold cylinder is a shallow depression termed the canyon, and another depression at each 2-fold axis is referred to as the dimple. Each icosahedral 3-fold axis is surrounded by a raised region (the 3-fold spike), which contains a number of overlapping antigenic sites (14, 15) as well as the binding sites of the host TfRs (16, 17).
TfR is a single-pass type-II membrane glycoprotein composed of 2 monomers joined by 2 disulfide bonds. TfR mediates iron uptake into cells by binding and importing iron-loaded transferrin (Tf), which may also bind to the HFE (hereditary hemochromatosis protein) in the intestine to regulate iron uptake by blocking Tf binding (reviewed in ref. 18). Structures of human TfR (hTfR) alone and of hTfR in complex with Tf have been solved by X-ray crystallography and by cryo-electron microscopy (cryo-EM) (19–25). Also, recently, high-resolution cryo-EM structures of TfR in complex with PvRBP2b (Plasmodium vivax reticulocyte-binding protein 2b) and ferritin were solved (26, 27). The TfR forms a butterfly-shaped dimeric ectodomain, with an about 30-Å-long stalk domain, a transmembrane sequence, and an N-terminal cytoplasmic tail that associates with AP2 (adaptor protein 2) to mediate uptake through clathrin-mediated endocytosis (28, 29). Each of the TfR ectodomain monomers comprises a protease-like domain that is closest to the membrane, a helical domain that also provides the dimeric interface, and a membrane-distal apical domain which does not have a known function in host biology. The 2-domain Tf molecules and the HFE protein bind primarily to the protease-like and helical domains of the dimeric TfR (25), while CPV capsids bind to the apical domain, as do the arenavirus glycoprotein (30), the envelope protein of mouse mammary tumor virus (31), ferritin (27), and the PvRBP2b (26).
In the current model of the control of host range, a series of key mutations in the capsid genes enabled CPV to adapt to the new canine host by allowing it to bind the canine TfR and use it as a functional receptor for entry and infection (32). Genetic mapping studies identified differences between FPV and CPV-2 that control canine host range as including VP2 residues 93 and 323 on the capsid surface (14, 16, 33). Introducing the CPV versions of those 2 residues into FPV conferred canine TfR binding and canine cell infection abilities (16). Other viral changes that both alter the viral host range and influence the binding of TfRs from different hosts include residues around VP2 residue 300 (9).
Mapping sites in the TfR that affect virus binding showed that the key residues were all within the apical domain of the receptor (34, 35). A key variation in the canine TfR was a Lys to Asn substitution of residue 384 (in the canine TfR sequence), which introduces a glycosylation site into the apical domain, and the added glycan blocked the binding of FPV and related viruses to the canine TfR (8, 34). Notably, TfR from bbj (Canis mesomelas) (bbj-TfR) is evolutionarily close to the canine TfR (98.6% amino acid sequence identity), but lacks the glycosylation site on residue 384. Bbj-TfR also binds CPV capsids, with both higher binding affinity and occupancy compared to the canine TfR (36). Thus, the CPV and bbj-TfR complex provides an ideal model for studying the molecular interactions between virus and receptor to understand how TfR binding controls viral host range.
The interactions between CPV and TfR are intricate, as there appears to be no direct relationship between the affinity of TfR binding and the success of the infection; indeed, some receptor mutants bind with high affinity but do not mediate infection, whereas the binding of the infection-competent canine TfR has very low affinity (36, 37). The capsids only engage 1 or a small number of TfRs on the surface of feline cells and also bind only small numbers of receptors when capsids are mixed with purified feline TfRs (17, 36). In the previous cryo-EM study using CPV and feline TfR, we predicted steric interference between adjacent TfR molecules and examined the TfR footprint as well as its binding mode, but precise interpretation was limited due to the low resolution of the asymmetric complex map that was possible at that time (17).
Here, we have solved the 3D structure of the CPV–TfR complex using cryo-EM single-particle analysis. The use of CPV and bbj-TfR provided a stable complex, while limiting the number of TfR molecules incubated per capsid ensured a large number of distinct virus–receptor complexes for analysis. Icosahedrally averaged reconstruction approaches did not reveal the TfR–capsid structure, but were used to solve the capsid structure at high resolution, as well as receptor-induced local conformational changes. Symmetry-mismatch reconstruction (SMR) was then used to show the receptor density on the capsid, and focused 3D classification increased the resolution of the asymmetric complex map and quantified the bound receptors. We also identified local variation of the bound receptors, which also induced the dynamic interaction of TfR binding to the capsid. Our proposed model therefore reveals the underlying mechanisms controlling cell binding and infection of different hosts by this virus, and thereby suggests how host-jumping emerging viruses can arise.
Results
Icosahedrally Averaged Reconstruction Reveals Enhanced Loop Flexibility Induced by Receptor Binding.
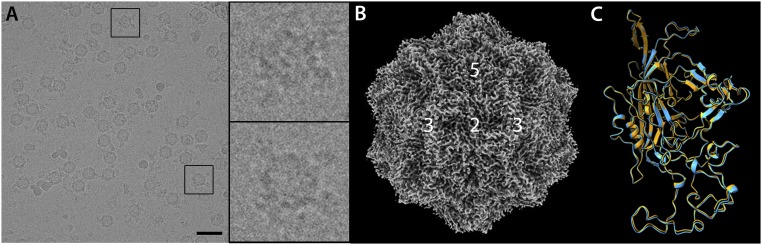
Previous work and the likely steric interference of bound adjacent TfR molecules suggested no more than 12 copies of the receptor bind per capsid (17, 36). Thus, the aim of this work was to solve an asymmetric reconstruction of complex. We used bbj-TfR for our structural studies to produce stable virus–receptor complexes. Purified CPV empty capsids were incubated with bbj-TfR ectodomains at a CPV:TfR ratio of 1:4, a ratio that successfully produces complexes while avoiding TfR-induced aggregation reported previously (17, 38). The incubated sample was applied to a grid, vitrified, and imaged on a Titan Krios with Falcon 3EC camera (Fig. 1A). Examination of the cryo-EM micrographs revealed capsids with extra densities, measuring 100–150 Å in length, consistent with the presence of CPV–TfR complexes (Fig. 1A, Insets). However, it was difficult to estimate the number of TfRs associated with each capsid by eye from the original micrographs.
Fig. 1.
Cryo-EM analysis of CPV in complex with TfR. (A) Representative image of the CPV–TfR complex micrographs. (A, Insets) The receptor density associated with the capsid structure was identified. (Scale bar, 50 nm.) (B) Surface-rendered map (gray) of the CPV capsid was reconstructed at 3.0-Å resolution using 60-fold symmetry averaging. The icosahedral symmetry axes were marked. (C) Superimposition of the VP2 structures for the capsid in the CPV–TfR complex (cyan) and the noncomplexed CPV capsid (PDB ID code 2CAS) (orange). VP2s are depicted as ribbon diagrams.
A single-particle reconstruction performed by using RELION 3.0 with icosahedral symmetry averaging resulted in a 3.0 Å resolution map (Fig. 1B). This map conformed to the canonical structure of CPV capsid, displaying the 5-fold cylinder with canyon, 3-fold spikes, and 2-fold dimple. An atomic model of the capsid structure was built into the icosahedral cryo-EM densities, and the overall rmsd between equivalent Cα atoms of the refined model and a CPV crystal structure was 0.552 Å (Fig. 1C) [Protein Data Bank (PDB) ID code 2CAS] (39). The refined atomic model revealed overall higher temperature-factor (or B-factor) distributions in the capsid surface compared to the CPV capsid alone (SI Appendix, Fig. S1). The 2 surface loops at the 5-fold cylinder and between the 2-fold and 3-fold symmetry axes showed the highest B factors in both structures. Notably, multiple surface loops comprising the 3-fold spikes in the receptor-associated capsid had B factors significantly higher than the rest of the capsid, which were not observed in the CPV capsid alone of any crystal structures we compared (12, 13, 33, 39–41). The local resolutions of the cryo-EM map agreed with the observed B factors (SI Appendix, Fig. S2). These findings suggest that the receptor binding induced conformational changes, specifically by increasing the flexibility of the loops on the 3-fold spikes, where TfR is known to bind (16, 17).
An Asymmetric Reconstruction Revealed the 3D Structure of Virus–Receptor Complex.
The icosahedrally averaged map of the capsid–TfR complexes did not resolve any cryo-EM densities corresponding to the receptor molecules, even at low contour levels. The absence of the receptor densities in the averaged 3D reconstruction was expected due to our incubation with only ∼4 TfRs per capsid. To resolve the TfRs bound to the capsid, we proceeded by applying a SMR method to solve an asymmetric structure of 1 receptor bound to the capsid (42–45).
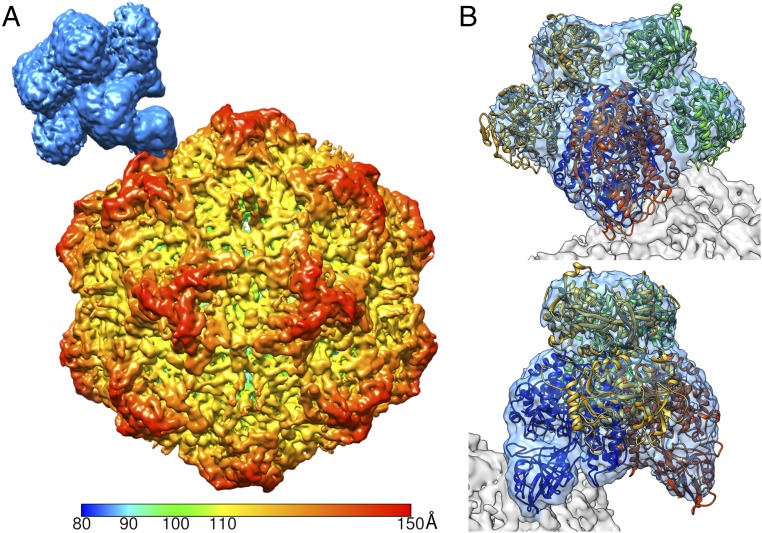
To begin the asymmetric structural analysis of the CPV–TfR complex, we made an initial model of a single hTfR molecule attached to the icosahedral map adjacent to the receptor binding site identified from the previous structural study (17). This model was used to perform an SMR, which selected 1 of the best-matching orientations for each particle image from the 60 orientations calculated from the previous icosahedrally averaged map. Although the TfR density was manually placed in the initial model, the obvious gradual changes throughout the early iterations during the SMR process showed that there was no model bias (SI Appendix, Fig. S3). After 2 rounds of multiple iterations of the SMR, an asymmetric map was reconstructed at 6.2-Å resolution which clearly showed the virus capsid with a putative receptor density attached near the tip of the 3-fold spike (Fig. 2A). Although the capsid density had distinct structural features, corresponding to 3- to 4-Å resolution, the receptor density was less well resolved.
Fig. 2.
SMR identifying the asymmetric structure of the CPV–TfR:Tf complex. (A) Surface rendering of the virus–receptor complex structure. The capsid was radially colored from the center according to the color key, whereas the receptor molecule was colored light blue. (B) Fitting of the TfR (blue and orange) and 2 Tfs (gold and green) into the cryo-EM density showed that the tip of the apical domain of TfR contacts the capsid surface. B, Upper and Lower represent 90° rotated views around the y axis.
An interpretation of the receptor density was initiated by fitting the structure of hTfR (PDB ID code 1CX8), which showed the hTfR dimer fitting properly; however, the receptor density had a significantly larger volume than that of hTfR alone (298,000 Å3 vs. 196,600 Å3). The fitted hTfR structure revealed that the volume differences were caused by 4 additional unfilled densities, located on each side of the TfR, adjacent to the protease-like and helical domains. These densities resembled 2 Tf molecules bound to the TfR dimer (20–22). When the TfR–Tf complex structure (PDB ID code 1SUV) was fitted into the density, the 2 unfilled densities on each side properly accommodated the 2 lobes of Tf structure with an overall correlation coefficient of 0.90 (Fig. 2B). Analysis of bbj TfR ectdomain preparations with charge detection mass spectrometry (CDMS) revealed that the TfR preparation used to make the receptor–virus complex contained masses consistent with TfR–Tf heteromers, where the TfRs were associated with either 1 or 2 Tfs (SI Appendix, Fig. S4).
The fitted TfR:Tf structures also revealed the nature of the steric collisions that would occur around each icosahedral 5-fold symmetry axis (Fig. 3A). The fitting showed that any 2 receptors located around a single 5-fold axis collided between TfR–TfR, TfR–Tf, or Tf–Tf. Steric collisions were identified only between the 5-fold–related TfR:Tf heteromers, so that this model indicated that up to 12 receptors, but likely no more, could bind to each capsid. Despite the quality of the fitting, the TfR:Tf cryo-EM density was not good enough to refine or build atomic coordinates for the entire TfR:Tf structure. Notably, the TfR monomer (mTfR) interacting with the virus surface had stronger density than the other portions of the receptor, including Tfs that were distal from the capsid (Fig. 2B). The local resolution map revealed poorer resolution that corresponded to the density variation in the asymmetric map (SI Appendix, Fig. S5). This finding suggested that not all TfRs had 2 Tf molecules bound, and there were conformational variations within the bound TfR molecules relative to the capsid binding site. To resolve the 2 issues and improve the receptor structure, we 1) increased the number of TfRs included in the 3D reconstruction by identifying the additional TfR-occupied sites on each capsid; 2) classified the newly found receptor densities; and 3) refined the receptor structure locally around the capsid binding site, as that is of greatest interest for understanding the structure and functions of the capsid–TfR interactions that control infection and host range.
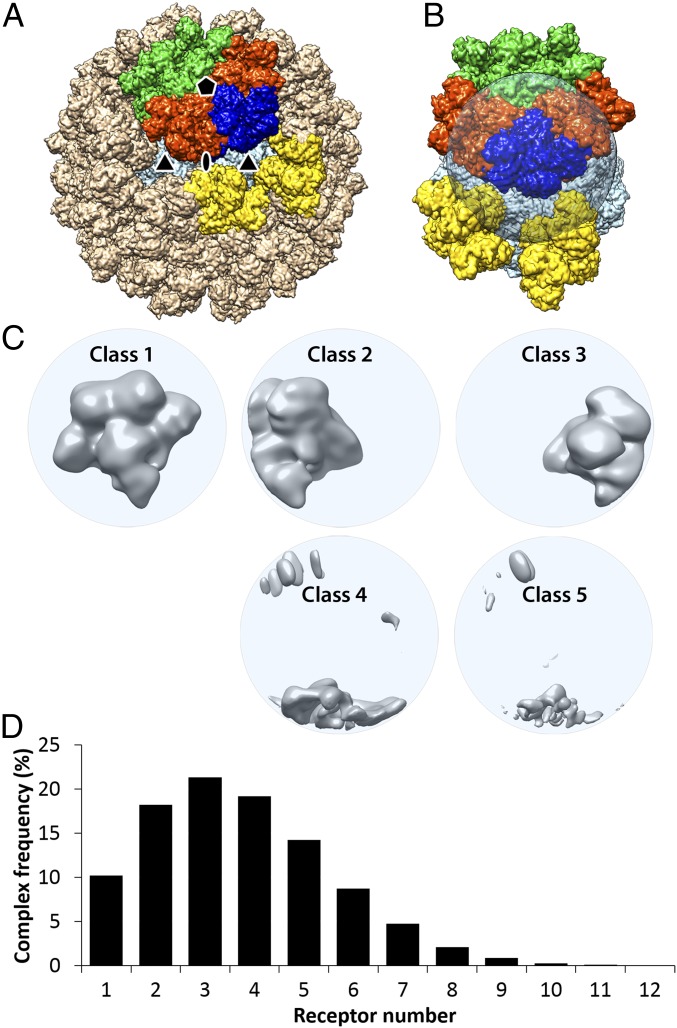
Fig. 3.
Focused 3D classification with symmetry-expanded orientation data characterizing the receptor binding sites based on the degree of receptor occupancy. (A) A simulation of 60 TfR:Tf heteromers bound to the CPV capsid was calculated by applying icosahedral symmetry operators to the bound TfR molecule in the symmetry-mismatch map (Fig. 2). The targeted volume occupied by the receptor was colored in blue; the 2 adjacent heteromers around the 5-fold axis were colored in orange; those across the symmetry axis were colored in green; those around the 3-fold symmetry axis were colored in yellow; and all other heteromers were colored in tan. (B) The transparent spherical mask applied for the target volume with occupying receptor colored in blue. (C) Five 3D-class reconstructions are shown in gray and surface rendered. The first class had full receptor densities. Classes 2 and 3 had partial densities corresponding to an unoccupied target volume plus the nearby orange receptors in A and B. The other classes had only some overlapping densities as shown. (D) A histogram for number of heteromers bound on each capsid. The average number of receptors bound to each capsid was 3.7.
Multiple Receptor Binding Sites Were Identified on the Capsid by Symmetry-Expanded Focused 3D Classification.
The SMR used only a single orientation for each capsid that positioned one of the bound receptor densities into the focused area. The objective of this approach was to reconstruct 1 receptor interacting with the capsid. Other bound TfR molecules, which were clearly seen in the original micrographs (Fig. 1A), were in random positions relative to the specific one oriented into the focused area for reconstruction. Thus, the small number of other TfRs on the capsid were averaged out with unoccupied binding sites outside the focused area in the SMR. Nevertheless, the asymmetric virus–receptor complex map identified the composition and location of the receptor precisely so that further structural analyses could be performed.
To separate the receptor-occupied binding sites from the unoccupied sites on the same capsid, we performed a focused 3D classification using a symmetry-expanded data file, which shows all 60 icosahedral orientations (Materials and Methods). Instead of choosing a single orientation for each particle, every particle was subjected to the 3D classification 60 times, using all 60 different orientations that are icosahedrally related. By applying a spherical mask at one of the receptor positions, multiple receptor densities on a single capsid could then be examined individually (Fig. 3B). The resulting cryo-EM reconstructions classified the receptor binding sites based on full, partial, or lack of the receptor densities (Fig. 3C). Among the 60 orientations, those that placed TfRs into the masked area were classified into the first class. Orientations that placed TfR-unoccupied binding sites at the masked area were classified into the other 4 classes. Two partial densities resulted from adjacent receptors around the 5-fold symmetry axis encroaching into the spherical mask (Fig. 3B, orange density). By identifying all of the receptor-occupied sites, we determined that 91.6% of the particles had at least 1 TfR:Tf heteromer bound. After discarding particle images that had no receptors bound or that had invading neighboring particles, the number of the bound receptors per capsid ranged from 1 to 12 with an average of 3.7 (Fig. 3D), close to the incubation condition of 4.
This classification scheme allowed us to identify a subclass of particles (6,237) that had only 1 receptor molecule bound on the capsid. Using this subpopulation, we generated an asymmetric map to examine the receptor-induced local-resolution changes on the 3-fold spikes. Although the quality of the map was not comparable with other maps, owing to the small number of particles, the local-resolution increase was observed on all the 3-fold spikes (SI Appendix, Fig. S6), suggesting that a single bound receptor induces flexibility to the 3-fold spike that propagates around the capsid. Although further examination of a subpopulation of capsids with only 2 TfRs showed a similar trend, a conclusion of receptor-induced long-range conformational change would need further examination.
A 3D Classification of the Receptor Density Resolved the Partial Occupancy of Tf Molecules and Revealed Conformational Variations of the TfR Structure.
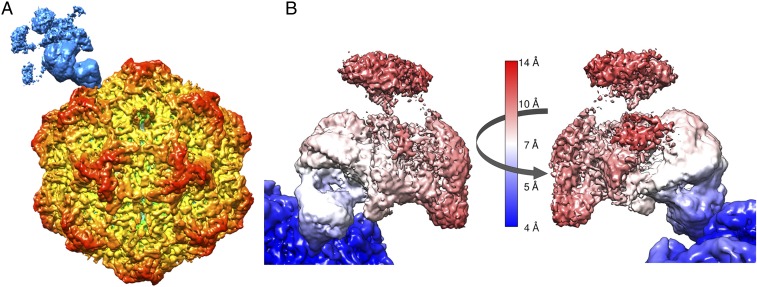
To improve the quality of the cryo-EM density of the receptor, the multiple orientations of each particle included in class 1 in Fig. 3C were used for a 3D reconstruction (Fig. 4A), thereby increasing the particle numbers by 2.6-fold (227,608 vs. 86,320). Compared to the asymmetric map using a single orientation per each particle, the global resolution of the complex map improved from 6.2 to 4.6 Å, as well as the local resolution of the receptor density (Fig. 4B). However, the feature of the receptor density changed. Specifically, the cryo-EM densities for the TfR monomer that was furthest from the capsid and its attached Tf molecule were significantly attenuated, whereas the density of the TfR monomer and the apical domain connected to the capsid remained similar to the previous result (Fig. 4B).
Fig. 4.
Asymmetric 3D reconstruction of the CPV–TfR:Tf complex using multiple orientations. (A) Surface-rendered 3D reconstruction using all orientations selected in Fig. 3C for each virus particle. The surface was colored according to the key as in Fig. 2. (B) Local resolutions of the receptor:Tf heteromer and adjacent capsid surface. Two orientation views of the receptor density (180° rotated) showed attenuation of the local density. The surface was colored according to the local resolution color key.
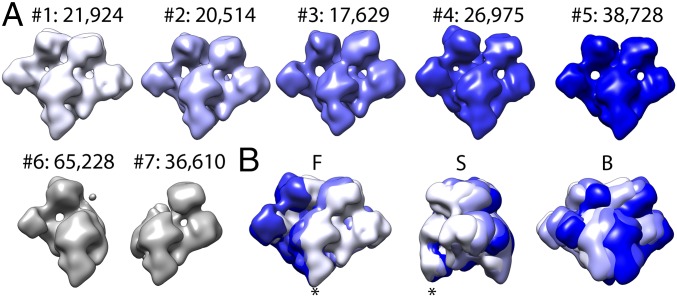
To understand why the TfR density appeared to weaken radially, we performed another round of 3D classification of the TfR:Tf densities into 7 distinguishable classes. The classification process was performed without particle alignment; therefore, the cryo-EM densities were classified based on the orientation of each receptor with respect to the capsid (Fig. 5A). The first result of this experiment was that 2 classes, comprising 45% of the total receptor population, had Tf density missing from one side of the TfR (Fig. 5A). Therefore, partial occupancy of Tf contributed to weaker TfR:Tf densities. Secondly, we found that all 7 resulting cryo-EM structures had different orientations with respect to the capsid, and none of them were superimposed perfectly (Fig. 5B). The local angular variation of the receptor structure appeared to be due to a small hinge motion previously identified between the TfR apical domain and protease-like domain (19), as well as from a pivoting motion of the TfR:Tf heteromer relative to its attachment site on the CPV capsid, which we investigated further after improving the TfR structure.
Fig. 5.
Focused 3D classification of the TfR:Tf cryo-EM density. (A) The 7 3D classes, where nos. 1–5 have 2 Tfs bound to the TfR, while in classes 6 and 7, only 1 Tf is bound. The numbers of each class collected from the complete dataset are indicated. (B) Superimposition of the 5 classes (nos. 1–5) are shown. B, back-side view; F, front view; S, right-side view.
Localized Reconstruction Improved the Quality of the TfR Structure.
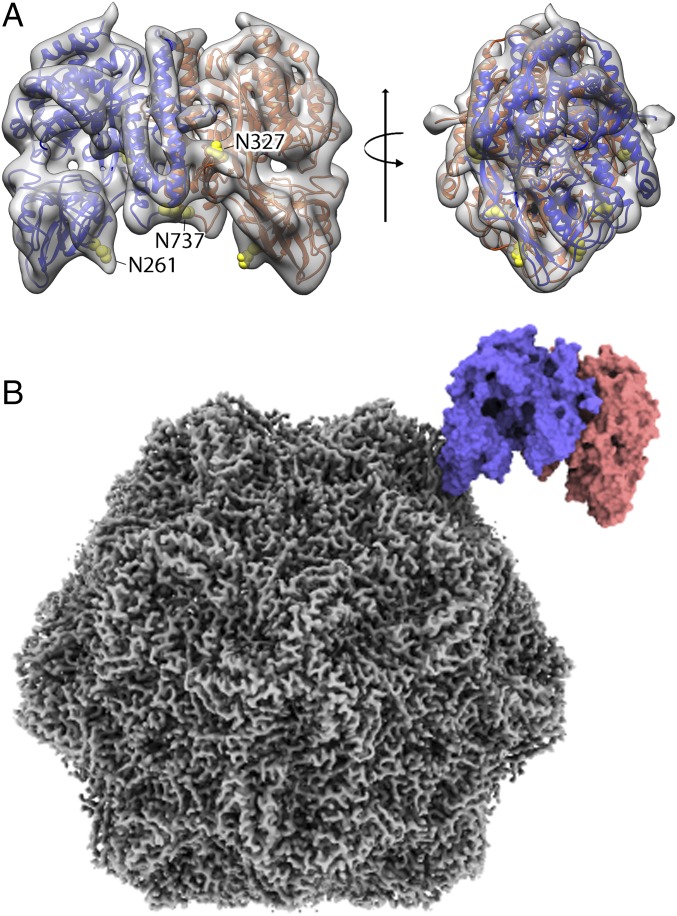
When 3D maps for each class were reconstructed as a whole virus–receptor complex, the receptor area still had locally varying resolutions, although the densities corresponding to the distal mTfR and bound Tf domains improved, compared to those in the previous reconstructions (SI Appendix, Fig. S7). This suggested that the variation of the bound receptor conformations was continuous relative to the capsid and could not be classified into a limited number of discrete states. Fitting the hTfR structure into the TfR density resulted in a poor fitting of the loop at the virus binding apical domain. To improve the receptor density and to obtain a more accurate molecular model of the bbj-TfR structure, we refined the cryo-EM receptor structure by applying an image-processing method, called localized reconstruction (46). The particle images were subtracted for the capsid densities, and then the receptor structure was iteratively refined with the local search option in RELION. The refined particle information produced a final map at 6.7 Å (Fig. 6A). The angular differences between the original and refined orientations of each particle showed a broad range of interactions, with a mean of 12° (SI Appendix, Fig. S8A), supporting the proposed rocking-motion model of the TfR relative to the capsid. A 3D classification of the TfR density revealed the 3 different populations as in Fig. 5; however, we included all TfRs together for the final map to increase the particle number. The new map resolved the entire structural features of the receptor and allowed refinement of the atomic model at a subnanometer resolution (Fig. 6A). Although side chains could not be defined without ambiguity due to the suboptimal resolution, the refined model corrected the virus-binding loop conformation and could be used for fitting to identify contacts (Fig. 6B). The TfR map also showed extra densities on the 3 glycosylation sites of the bbj-TfR, residues 261, 327, and 737, at a contour level lower than 2σ.
Fig. 6.
The localized reconstruction of the receptor density and fitted model identifying the TfR footprint on the CPV surface. (A) A 6.7-Å resolution cryo-EM density map for the bbj-TfR was generated by localized reconstruction image processing without symmetry. The intermediate resolution map was used to refine the bbj-TfR atomic model without the Tf bound. The TfR 3D map and model (with monomers colored in blue and red ribbons) were superimposed (A, Left) and rotated by 90° (A, Right). Three glycosylation sites with extra densities were shown by atoms in yellow spheres and labeled (A, Left). The local 3D map was surface rendered and colored transparent gray. The Tf density was removed to allow visualization. (B) The refined TfR model fitted into the asymmetric complex map. The sharpened map for CPV capsid (gray) and the fitted TfR model (blue and red) was surface rendered.
Morph Analysis and Normal Mode Analysis Revealed Swaying Motion of the Bound Receptor.
The refined TfR structure was fitted into the 5 subclasses in Fig. 5, with correlation coefficients higher than 0.94. Using the fitted TfR structures, we also generated a morph movie that showed the rocking motion of the bound receptor in which the whole receptor molecule swayed relative to the capsid, with the binding site acting as a pivot point (Movies S1 and S2). The 2 most distant TfR:Tf positions differed by about 20 Å at the center of the complex and about a 16° angle between axes of the molecules (SI Appendix, Fig. S8B). This wide-range rocking motion of the TfR cannot be explained by a simple hinge motion between the apical and protease-like domains, but more likely is due to movement of the large (450-kDa) TfR:Tf heteromeric complex relative to the small attachment footprint near the tip of the TfR apical domain.
To simulate the rocking motion, we performed normal mode analysis (NMA) for the CPV–TfR:Tf complex structure with Cα-based coarse-graining model by using iMOD (47). The 3 low-frequency normal modes (modes 1, 2, and 3) (Movies S3, S4, and S5) described the largest movements in a protein and included more than 90% of the variance (SI Appendix, Fig. S9). Mode 1 corresponded to the rocking motion along the primary direction of the morph analysis, whereas modes 2 and 3 corresponded to the rocking motion normal to the mode 1 and rotation around an axis normal to the capsid surface, respectively. The motions observed in morph analysis could be explained by these 3 modes. As the global modes of NMA are dominated by the overall shape, form, or native contact topology of the protein and are insensitive to local interactions (48), the swaying motion was caused by concerted domain movements of the 2 large molecules (CPV and TfR:Tf), interacting through a small contact area.
Dynamic Interactions between CPV and TfR.
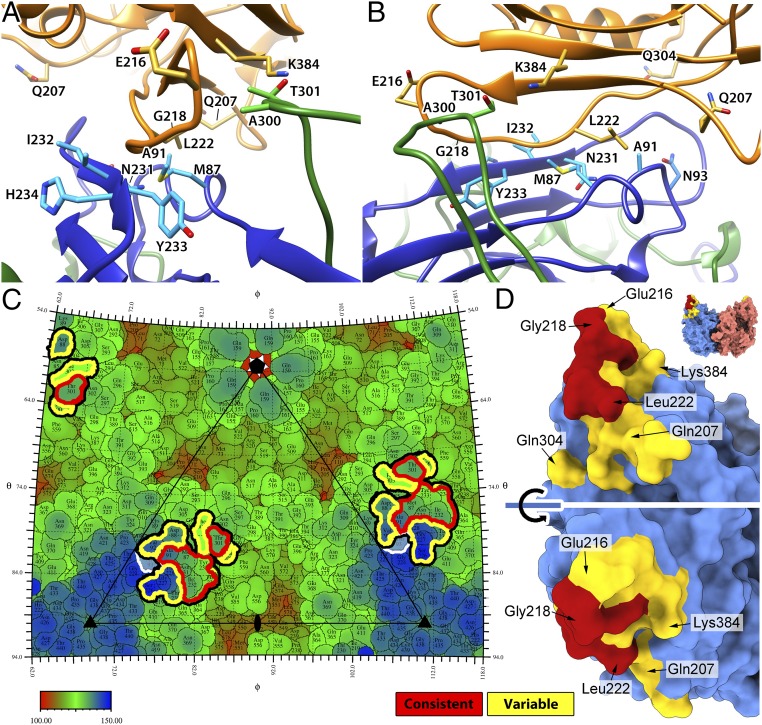
The swaying movement of the receptor on the capsid surface implied variation of the virus–receptor interactions in different 3D classes. The fitted models also showed significantly different interatomic clashes at the CPV–TfR interface (ranging from 21 to 45 pairs), suggesting local conformational changes on both structures. The interactions between CPV and TfR identified from the 5 classes showed a dynamic interface. The residues found in all 5 models were designated as “consistent” and all of the other residues as “variable,” although the terminology does not imply biological significance of the residues (Table 1). The receptor-contacted residues on the capsid were all located within the BC, EF, and GH loops, near the tip of the 3-fold spike (Fig. 7 A–C). Those 3 loops were intertwined around the 3-fold axis, with the BC and EF loops contributed by the same copy of a capsid protein and the GH loop from a different, symmetry-related capsid protein. The BC and EF loops appeared to have consistent interactions with the TfR (identified from all 5 classes), as well as variable interactions (identified in from 1 to 4 classes), while the GH loop had only consistent interactions (Table 1). The EF loop contained more than half of the contacting residues (10 of 17), including 4 consistent interactions on a short β-strand (β″EF, residues 231–234) and 6 variable interactions that may not be used in every case (Table 1). Among the other interactions were those with residues Met-87 (consistent, BC loop) and Ala-300 (variable, GH loop), which have showed clear host-specific evolution in CPV since its emergence in dogs (9). Most of the variably interacting sites have not been identified previously in studies of receptor binding or of viral evolution.
Table 1.
Contacting residues identified from the fitting of TfR into the 5 3D classes
| Location | Residue | No. 1 | No. 2 | No. 3 | No. 4 | No. 5 | Count |
| CPV chain-1 | |||||||
| BC loop (αBC: 86–91) | Met-87 | O | O | O | O | O | 5 |
| Asp-88 | O | O | 2 | ||||
| Ala-91 | O | O | O | O | O | 5 | |
| Val-92 | O | 1 | |||||
| EF loop (β’’EF:230–235) | Ser-226 | O | O | 2 | |||
| Gly-227 | O | O | O | O | 4 | ||
| Thr-228 | O | O | O | O | 4 | ||
| Pro-229 | O | O | O | O | 4 | ||
| Thr-230 | O | O | O | O | 4 | ||
| Asn-231 | O | O | O | O | O | 5 | |
| Ile-232 | O | O | O | O | O | 5 | |
| Tyr-233 | O | O | O | O | O | 5 | |
| His-234 | O | O | O | O | O | 5 | |
| Gly-235 | O | O | 2 | ||||
| CPV chain-2 | |||||||
| GH loop | Ala-300 | O | O | O | O | 4 | |
| Thr-301 | O | O | O | O | O | 5 | |
| Phe-303 | O | 1 | |||||
| No. of residues | 17 | 13 | 11 | 13 | 15 | 11 | |
| TfR | |||||||
| βI-1/βII-1 loop | Asn-205 | O | 1 | ||||
| Gln-207 | O | 1 | |||||
| βII-1 | Thr-209 | O | 1 | ||||
| Thr-211 | O | O | 2 | ||||
| Val-213 | O | O | O | O | O | 5 | |
| βII-1/βII-2 loop | Gly-214 | O | O | 2 | |||
| Met-215 | O | 1 | |||||
| Glu-216 | O | 1 | |||||
| Ser-217 | O | O | O | O | O | 5 | |
| Gly-218 | O | O | O | O | O | 5 | |
| Leu-219 | O | O | O | O | O | 5 | |
| Val-220 | O | O | O | O | O | 5 | |
| Tyr-221 | O | O | O | O | O | 5 | |
| βII-2 | Leu-222 | O | O | O | O | O | 5 |
| Glu-224 | O | O | 2 | ||||
| Ser-225 | O | O | 2 | ||||
| Glu-227 | O | 1 | |||||
| βII-3 | Thr-239 | O | O | 2 | |||
| βII-6/βII-7 loop | Gln-304 | O | O | O | 3 | ||
| βII-8 | Asn-382 | O | O | 2 | |||
| Lys-384 | O | O | 2 | ||||
| No. of residues | 21 | 10 | 9 | 9 | 18 | 12 |
The contacting residues on each surface (2 CPV chains and TfR) were listed and marked “O” for each class shown in Fig. 5. The residues were grouped for their secondary structures. Residues found in all 5 classes were denoted as “consistent” interactions and residues found in less than 5 classes were denoted as “variable” interactions.
Fig. 7.
The fitting of the TfR structure identifies the dynamic interactions between CPV and TfR. (A and B) The refined bbj-TfR structure was fitted into the 5 cryo-EM maps. Fitted structures for class 3 are depicted. The 2 CPV VP2 chains and TfR are shown as ribbon diagrams and colored in blue, green, and orange, respectively. Side chains of the consistent contacts as well as Asn-93 and Ala-300 of CPV residues and TfR residues 216, 218, 207, 222, 304, and 384 are shown as stick models and labeled. (C) The roadmap of the CPV capsid surface with TfR footprint indicated. All of the receptor-contacting residues are highlighted by thick black lines. The consistent and variable contacts are further highlighted in red and yellow lines, respectively. CPV residue 93 is marked by white lines. The capsid was radially colored according to the color map, and 1 asymmetric unit is indicated by a triangle with icosahedral symmetry axis marks. (D) The capsid-contacting residues on the model of the bbj-TfR molecule. The TfR was surface rendered, and each monomer was colored differently (top). The residues that are seen to make consistent and variable contacts with the capsid were colored red and yellow, respectively. The large red surface area includes residues from the βII-1/βII-2 loop in addition to Val-213 and Leu-222.
The TfR surface that interacted with the CPV capsid involved the tip of the apical domain, including multiple β-strands and loops (Fig. 7D and Table 1). Most consistent TfR interactions were located on the βII-1/βII-2 loop, including residues 217–221 as well as Leu-222, a key residue for CPV binding (34) (Fig. 7D). These residues contacted most of the consistently interacting CPV residues from all 3 capsid loops. TfR residue Gly-218, along with 3 variably interacting residues, Gln-207, Glu-216, and Gln-304, has been identified previously as being under positive selection (Table 1) (8). Notably, Lys-384 on the bbj-TfR was in contact with VP2 residue Thr-301 on CPV—in canine TfR, this was replaced by Asn-384, which forms the unique glycosylation site. VP2 residue Thr-301 has also been associated with host-specific variation and is located on the CPV surface to one side of the TfR binding area, with the other side including the EF loop that was associated with the large variable interactions (Fig. 7C).
Binding Analysis Showed TfR:Tf Binds Virus Better than TfR Alone.
The finding of the consistent occupation of the Tf binding sites was unexpected, and was likely due to the TfR associating with bovine Tf from the serum-free medium used for expression of the secreted TfR apical domains used in these studies. The Tf was not detected prior to the structural studies because it ran at the same position in sodium dodecyl sulfate/polyacrylamide gels as the TfR ectodomain monomer. To examine the effects of Tf binding to the TfR on the virus–receptor interaction, we used a bio-layer interferometry approach that has been detailed in a recent publication (4). Specifically, we tested capsid binding to the bbj-TfR that had Tf bound and to the bbj-TfR with Tf removed by low pH and iron removal. The results showed that the Tf-free TfR bound virus at a lower apparent affinity (or avidity) compared to the original Tf-complexed receptor (SI Appendix, Fig. S10), suggesting that Tf binding to the TfR enhances receptor binding to the capsid. Although we cannot exclude the possibility that the treatment to remove the Tf altered the capsid-binding properties of the receptor in the binding assay, a preference of virus for TfR–Tf was seen in the structural analysis.
Discussion
TfR Binding May Exert a Lever Force on the Capsid.
Our results showed that the small contact area between CPV and the single apical domain of the large TfR:Tf heteromer resulted in swaying of the mass of the TfR:Tf relative to the capsid surface, which, in turn, resulted in the dynamic interaction at the binding site. A mechanical force generated by the swaying motion may also induce the conformational changes of the 3-fold spikes that were observed as the decrease in local resolution and increase in temperature factor. Similar changes have been seen for complexes of CPV capsids with Fab E molecules, where the increased local disorder was interpreted as the neutralization mechanism for Fab E by preventing TfR binding (49). In the case of CPV–Fab E complexes, the capsids were saturated with Fab molecules; however, here we found that a single bound receptor induced capsid-wide flexibility (SI Appendix, Figs. S1, S2, and S6). The area with high temperature factors or lower local resolution was not limited to the TfR footprint, but included broader areas related by 3-fold symmetry, suggesting that the TfR binding-induced conformational changes might be propagated to the neighboring unoccupied binding sites. These changes initiated by receptor binding neither inhibit nor enhance subsequent TfR binding events on the capsid. Since parvovirus capsids are known to be highly stable, perhaps receptor-induced flexibility provides a function during the next steps in the entry process, although the transfer of inertia by a receptor binding needs further confirmation.
The Dynamic CPV–TfR Interaction Suggests a Rock-and-Roll Model.
The TfR footprints that were consistently found within all 5 3D classes included residues on 3 capsid loops. The footprint and immediately adjacent capsid structure contained multiple residues previously identified as controlling the virus–receptor interaction, CPV host-range determination, and host adaptation, as well as residues that have become widespread around the world during the evolution of the virus. These include VP2 residues 87, 232, 300, and 301 (9, 10, 50, 51), confirming that the TfR–capsid interactions control the host ranges of the virus and are likely driving much of the evolution of the capsid structure in this region. The TfR footprint also included multiple residues that have not been observed to be involved in host-range control.
The dynamic changes of the receptor footprints included residues with consistent and variable interactions (Fig. 7 and Table 1). The consistent interactions provided a pivot point, while the variable interactions supported the dynamic interaction, which resulted in the rock-and-roll motion of the 2 molecules. It should be noted that multiple alanine substitutions within the BC and GH loops of CPV, including some involved in consistent interactions (M87 and T301), did not cause loss of binding affinity to the bbj-TfR (36). Although the CPV residues responsible for binding the bbj-TfR are not known, the tolerance of the Ala substitutions provides useful information. Based on these observations, we propose a model where the multiloop and dynamic interaction allows more viral host range variation, as it would permit some unfavorable changes to be tolerated on the capsid surface during the process of virus adaptation to the TfR of a new host. This buffering effect would also allow the virus to pass across evolutionary valleys between the fitness peaks of optimized interactions with the TfRs from different hosts (52).
The Dynamic Interaction Explains the Molecular Mechanism for CPV Host Jump to Dogs.
The additional glycosylation at the residue 384 of the canine TfR would have introduced a high barrier preventing the host jump of the ancestor of CPV into dogs (32). Multiple factors, including the flexibility in the binding site and an efficient infection by CPV, likely allowed the virus to become adapted to the glycosylated canine TfR, permitting CPV to emerge and cause the pandemic of canine disease.
Our model of the CPV–TfR complex structure shows the bbj-TfR residue K384 (equivalent to N384 of the canine TfR) interacting with CPV residue Thr-301 on the GH loop (Fig. 7B). Attachment of the large glycan ((Hex)2(HexNAc)1(NeuAc)1 + (Man)3(GlcNAc)2: 1,710.6 Da (31)) to this residue in canine-TfR would likely push the TfR away from that loop. This, in turn, would increase the proximity to the dynamic interaction site on the EF loop, located on the opposite side from GH loop within the footprint. In that region, the variably interacting residues on the EF loop (VP2 residues 226–230) are adjacent to VP2 residue 93, a key residue for canine host-range determination (13, 15). In contrast to FPV, which has Lys at 93 that forms hydrogen bonds with both residues 225 and 227, CPV contains Asn-93 and lacks both hydrogen bonds (30). The increased flexibility of the EF loop is also supported by the increased temperature factors of those residues after the receptor binding. Another flexibility introduced into the GH loop during the emergence of the CPV-2a strain resulted from the Ala–Gly substitution of VP2 residue 300. Notably, the canine TfR supports the high infectivity of CPV, despite the low binding affinity to canine TfR (34). It appears that a rapid uptake of the viruses from cell surface (25) may allow infection, despite the weak canine TfR:capsid interactions. In addition, our data showing the higher affinity of TfR:Tf heteromer to CPV suggest that the virus may hijack the iron-loaded TfR:Tf internalization for efficient virus infection. The combined effects of the enhanced dynamic interaction with increased flexibility of the receptor binding loops and the weak TfR interaction of CPV counterbalanced by the efficient virus infection would have permitted cross-species transmission of CPV.
The dynamic virus–receptor interaction as a driving evolutionary force raises questions about the possibility of a CPV species jump into additional hosts, including humans. Another important point is whether this dynamic interaction between virus and host receptor affects cross-species transmission of other viruses. Our additional normal-mode analyses on complex structures of Machupo virus GP1 complexed with TfR (PDB ID code 3KAS) (53) showed similar motions as in the CPV–TfR complex, but at a different magnitude. Multiple factors would affect the dynamic interactions, such as the topology of the contacts, binding affinity, or molecular size of the virus and receptor. Detailed structural analysis along with molecular dynamics and genetic information is needed to confirm the general application of the rock-and-roll model for other viruses.
Atomic resolution maps of the virus–receptor complex structures are critical to understand the molecular mechanisms that have not been seen previously. Our results provide important interpretations of previous evolutionary and mutational studies that revealed the virus and receptor sequences necessary and sufficient for receptor recognition. These results also explain how the key interactions between variant viruses and different host TfRs control virus binding to cells leading to infection and how the viruses may evolve to gain new host ranges.
Materials and Methods
Virus Purification and Protein Preparation.
CPV empty capsids were purified from infected cells via density gradient centrifugation, as described (36, 37). Briefly, feline kidney cells were infected, and virus capsids were purified from lysates by sucrose gradient centrifugation. Soluble, His-tagged bbj-TfR was expressed in BHK cells, purified as described (36), and used within 4 h of purification from the culture medium.
Cryo-EM and Data Collection.
CPV (1.52 mg/mL) was incubated with bbj-TfR (0.61 mg/mL) for a ratio of 1:4 at room temperature for ∼30 min. For cryo-grid preparation, a 3-µL aliquot of the incubated sample was applied to a fresh glow-discharged QUANTIFOIL EM grid. The grids were blotted for 3.5 s in 95% relative humidity for plunge-freezing (Vitrobot; Thermo Fisher) in liquid ethane. Cryo-EM datasets were collected at 300 kV with a Titan Krios microscope (Thermo Fisher), equipped with a spherical aberration corrector. Movies (linear mode: 15 frames, each 0.12 s, total dose 150 e−Å−2) were recorded by using a Falcon 3 detector with a defocus range of 0.4–4.4 µm. Automated single-particle data acquisition was performed with EPU with a nominal magnification of 59,000×, which yields a final pixel size of 1.11 Å/pixel. Data-collection parameters and dataset size are shown in SI Appendix, Table S1.
Image Processing.
A total of 3,726 movies were recorded. Frame alignment and dose weighting were performed with MotionCor2 (54), and contrast transfer function (CTF) parameters were estimated with GCTF (55). Out of these, 3,366 micrographs with CTF figure-of-merit greater than 0.1 were selected. Initially, particles were picked and extracted in RELION (Version 2) with 400 × 400 pixels (56). After 2D classification, ∼154,000 particle images were selected and used for a 3.0-Å (Fourier shell correlation = 0.143) resolution icosahedral reconstruction. Then, RELION (Version 3.0-beta) was used for local CTF refinement and Bayesian polishing (57). The resulting shiny particles were used for the final-resolution icosahedral refinement and postprocess, resulting in a 3.06-Å resolution map.
SMRs.
The SMR process was performed as described with our modified version of RELION (Version 3.0) (42). Briefly, the icosahedrally refined particles were reextracted with box size of 512 × 512 pixels and binned by factor 2. For a starting model, a density map of the CPV–TfR complex was generated in Chimera (58) by placing hTfR ectodomain (PDB ID code 1CX8) on a CPV capsid map across the 2-fold symmetry axis so that the each apical domain touched the 2 3-fold spikes mirror-positioned across the 5F–2F symmetry axis line. The initial map was low-pass-filtered at 60 Å prior to SMR. Particle center and orientations from the icosahedral 3D refinement were used for SMR. The custom option flag “--break_symmetry” was used with the relion_refine_mp program to select the best-matching single orientation among the 60 icosahedrally related orientations. Two rounds of 3D classification were performed to discard particles without receptor density or with nonsensical density. Two additional rounds of 3D classification with a single class were performed to refine the model.
Focused 3D Classification.
For this process, 2 × 2 binned images were used. The capsid densities were subtracted from particle images in RELION. Then, the 60 icosahedral symmetry-related orientations of each of the 62,005 particles selected from the SMR 3D classification were calculated by the RELION command “relion_particle_symmetry_expand.” The expanded STAR file, including 3,720,300 orientations, was subjected to 3D classification, using C1 symmetry without alignment of particles and with a spherical mask 180 Å in diameter at one of the receptor positions. The first round of the 3D classification identified the receptor-occupied and -unoccupied binding sites, selecting 227,608 orientations that located receptor density at the masked region. Second-round 3D classification using the selected orientations classified particles into 7 classes, sorting them based on Tf occupancy and orientations of the bound receptors relative to each other class.
Localized Reconstructions.
To reconstruct the receptor structure, particle images were reextracted with a box size of 580 × 580 pixels. “Relion_localized_reconstruction” in scipion package (46, 59) was used to extract subparticle images with a box size of 200 × 200. The subparticles were refined in RELION with local angular search mode, resulting in a 6.9-Å resolution map. The refined map was postprocessed at 6.7-Å resolution.
Homology Model.
To generate a homology model of the bbjTfR, the amino acid sequence of the bbj-TfR was aligned against that of the hTfR ectodomain (PDB ID code 1CX8), and then the aligned sequences were submitted to the automated protein structure homology-modeling server SWISS-MODEL (60).
Atomic Model Refinement.
The fitted structure of the CPV asymmetric unit (PDB ID code 2CAS) (39) was duplicated for 60 icosahedral units and refined with noncrystallographic symmetry using PHENIX real space refine (61). The map cross-correlation values for “whole unit cell” and “around atoms” were 0.748 and 0.832, respectively. The rmsd values for bonds and angles were 0.01 and 0.74, respectively. The Ramachandran outlier was 0.12%. The refined model was visually inspected in Coot (62) and validated by MolProbity (63).
Visualization and Statistics.
Map visualization and images were generated in Chimera (58) and ChimeraX (64). The capsid roadmap was generated by using RIVEM (65). Counting the number of the bound receptors in focused 3D classification was done by using R.
Supplementary Material
Acknowledgments
We thank Wendy Weichert for expert technical support; and Carmen Dunbar and Martin Jarrold for performing CDMS analysis. This work was supported by NIH Grants R01 AI092571 (to C.R.P. and S.L.H.) and R01 GM080533 (to C.R.P.). H.M.C. was supported by NSF Graduate Research Fellowships Program Grant DGE-1650441. This work was supported in part by the Pennsylvania Department of Health Commonwealth Universal Research Enhancement Program. Research reported in this publication was also supported by the Office of the Director, NIH, Award S10OD011986. The funders had no role in study design, data collection and interpretation, or decision to submit the work for publication.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.J.K. is a guest editor invited by the Editorial Board.
Data deposition: Data have been deposited in the Electron Microscopy Data Bank, https://www.ebi.ac.uk/pdbe/emdb/ (accession nos. EMDB-20001–20003) and the Protein Data Bank, https://www.rcsb.org/ (PDB ID code 6OAS).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904918116/-/DCSupplemental.
References
- 1.Geoghegan J. L., Duchêne S., Holmes E. C., Comparative analysis estimates the relative frequencies of co-divergence and cross-species transmission within viral families. PLoS Pathog. 13, e1006215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parrish C. R., et al. , Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol. Mol. Biol. Rev. 72, 457–470 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hueffer K., Parrish C. R., Parvovirus host range, cell tropism and evolution. Curr. Opin. Microbiol. 6, 392–398 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Parrish C. R., Kawaoka Y., The origins of new pandemic viruses: The acquisition of new host ranges by canine parvovirus and influenza A viruses. Annu. Rev. Microbiol. 59, 553–586 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Truyen U., et al. , Evolution of the feline-subgroup parvoviruses and the control of canine host range in vivo. J. Virol. 69, 4702–4710 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stucker K. M., et al. , The role of evolutionary intermediates in the host adaptation of canine parvovirus. J. Virol. 86, 1514–1521 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoelzer K., Parrish C. R., The emergence of parvoviruses of carnivores. Vet. Res. 41, 39 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaelber J. T., et al. , Evolutionary reconstructions of the transferrin receptor of Caniforms supports canine parvovirus being a re-emerged and not a novel pathogen in dogs. PLoS Pathog. 8, e1002666 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allison A. B., et al. , Single mutations in the VP2 300 loop region of the three-fold spike of the carnivore parvovirus capsid can determine host range. J. Virol. 90, 753–767 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allison A. B., et al. , Host-specific parvovirus evolution in nature is recapitulated by in vitro adaptation to different carnivore species. PLoS Pathog. 10, e1004475 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsao J., et al. , The three-dimensional structure of canine parvovirus and its functional implications. Science 251, 1456–1464 (1991). [DOI] [PubMed] [Google Scholar]
- 12.Wu H., Rossmann M. G., The canine parvovirus empty capsid structure. J. Mol. Biol. 233, 231–244 (1993). [DOI] [PubMed] [Google Scholar]
- 13.Xie Q., Chapman M. S., Canine parvovirus capsid structure, analyzed at 2.9 A resolution. J. Mol. Biol. 264, 497–520 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Chang S. F., Sgro J. Y., Parrish C. R., Multiple amino acids in the capsid structure of canine parvovirus coordinately determine the canine host range and specific antigenic and hemagglutination properties. J. Virol. 66, 6858–6867 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hafenstein S., et al. , Structural comparison of different antibodies interacting with parvovirus capsids. J. Virol. 83, 5556–5566 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hueffer K., Govindasamy L., Agbandje-McKenna M., Parrish C. R., Combinations of two capsid regions controlling canine host range determine canine transferrin receptor binding by canine and feline parvoviruses. J. Virol. 77, 10099–10105 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hafenstein S., et al. , Asymmetric binding of transferrin receptor to parvovirus capsids. Proc. Natl. Acad. Sci. U.S.A. 104, 6585–6589 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aisen P., Transferrin receptor 1. Int. J. Biochem. Cell Biol. 36, 2137–2143 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Lawrence C. M., et al. , Crystal structure of the ectodomain of human transferrin receptor. Science 286, 779–782 (1999). [DOI] [PubMed] [Google Scholar]
- 20.Cheng Y., Zak O., Aisen P., Harrison S. C., Walz T., Structure of the human transferrin receptor-transferrin complex. Cell 116, 565–576 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Cheng Y., Zak O., Aisen P., Harrison S. C., Walz T., Single particle reconstruction of the human apo-transferrin-transferrin receptor complex. J. Struct. Biol. 152, 204–210 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Cheng Y., et al. , Single particle reconstructions of the transferrin-transferrin receptor complex obtained with different specimen preparation techniques. J. Mol. Biol. 355, 1048–1065 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Fuchs H., Lücken U., Tauber R., Engel A., Gessner R., Structural model of phospholipid-reconstituted human transferrin receptor derived by electron microscopy. Structure 6, 1235–1243 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Bennett M. J., Lebrón J. A., Bjorkman P. J., Crystal structure of the hereditary haemochromatosis protein HFE complexed with transferrin receptor. Nature 403, 46–53 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Eckenroth B. E., Steere A. N., Chasteen N. D., Everse S. J., Mason A. B., How the binding of human transferrin primes the transferrin receptor potentiating iron release at endosomal pH. Proc. Natl. Acad. Sci. U.S.A. 108, 13089–13094 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruszczyk J., et al. , Cryo-EM structure of an essential Plasmodium vivax invasion complex. Nature 559, 135–139 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Montemiglio L. C., et al. , Cryo-EM structure of the human ferritin-transferrin receptor 1 complex. Nat. Commun. 10, 1121 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cureton D. K., Harbison C. E., Cocucci E., Parrish C. R., Kirchhausen T., Limited transferrin receptor clustering allows rapid diffusion of canine parvovirus into clathrin endocytic structures. J. Virol. 86, 5330–5340 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pérez M. J., Fernandez N., Pasquini J. M., Oligodendrocyte differentiation and signaling after transferrin internalization: A mechanism of action. Exp. Neurol. 248, 262–274 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Radoshitzky S. R., et al. , Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature 446, 92–96 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y., Rassa J. C., deObaldia M. E., Albritton L. M., Ross S. R., Identification of the receptor binding domain of the mouse mammary tumor virus envelope protein. J. Virol. 77, 10468–10478 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shackelton L. A., Parrish C. R., Truyen U., Holmes E. C., High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc. Natl. Acad. Sci. U.S.A. 102, 379–384 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Govindasamy L., Hueffer K., Parrish C. R., Agbandje-McKenna M., Structures of host range-controlling regions of the capsids of canine and feline parvoviruses and mutants. J. Virol. 77, 12211–12221 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palermo L. M., Hueffer K., Parrish C. R., Residues in the apical domain of the feline and canine transferrin receptors control host-specific binding and cell infection of canine and feline parvoviruses. J. Virol. 77, 8915–8923 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodman L. B., et al. , Binding site on the transferrin receptor for the parvovirus capsid and effects of altered affinity on cell uptake and infection. J. Virol. 84, 4969–4978 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Callaway H. M., et al. , Complex and dynamic interactions between parvovirus capsids, transferrin receptors and antibodies control cell infection and host range. J. Virol. 92, e00460-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Callaway H. M., et al. , Parvovirus capsid structures required for infection: Mutations controlling receptor recognition and protease cleavages. J. Virol. 91, e01871-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palermo L. M., Hafenstein S. L., Parrish C. R., Purified feline and canine transferrin receptors reveal complex interactions with the capsids of canine and feline parvoviruses that correspond to their host ranges. J. Virol. 80, 8482–8492 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simpson A. A., et al. , Host range and variability of calcium binding by surface loops in the capsids of canine and feline parvoviruses. J. Mol. Biol. 300, 597–610 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Llamas-Saiz A. L., et al. , Structural analysis of a mutation in canine parvovirus which controls antigenicity and host range. Virology 225, 65–71 (1996). [DOI] [PubMed] [Google Scholar]
- 41.Organtini L. J., Allison A. B., Lukk T., Parrish C. R., Hafenstein S., Global displacement of canine parvovirus by a host-adapted variant: Structural comparison between pandemic viruses with distinct host ranges. J. Virol. 89, 1909–1912 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee H., et al. , The novel asymmetric entry intermediate of a picornavirus captured with nanodiscs. Sci. Adv. 2, e1501929 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li X., Liu H., Cheng L., Symmetry-mismatch reconstruction of genomes and associated proteins within icosahedral viruses using cryo-EM. Biophys. Rep. 2, 25–32 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X., et al. , In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus. Nature 527, 531–534 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ding K., Nguyen L., Zhou Z. H., In situ structures of the polymerase complex and RNA genome show how aquareovirus transcription machineries respond to uncoating. J. Virol. 92, e00774-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ilca S. L., et al. , Localized reconstruction of subunits from electron cryomicroscopy images of macromolecular complexes. Nat. Commun. 6, 8843 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lopéz-Blanco J. R., Garzón J. I., Chacón P., iMod: Multipurpose normal mode analysis in internal coordinates. Bioinformatics 27, 2843–2850 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Bahar I., Lezon T. R., Bakan A., Shrivastava I. H., Normal mode analysis of biomolecular structures: Functional mechanisms of membrane proteins. Chem. Rev. 110, 1463–1497 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Organtini L. J., et al. , The near-atomic resolution structure of a highly neutralizing Fab bound to canine parvovirus. J. Virol. 90, 9733–9742 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parker J. S., Parrish C. R., Canine parvovirus host range is determined by the specific conformation of an additional region of the capsid. J. Virol. 71, 9214–9222 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parrish C. R., Host range relationships and the evolution of canine parvovirus. Vet. Microbiol. 69, 29–40 (1999). [DOI] [PubMed] [Google Scholar]
- 52.Geoghegan J. L., Senior A. M., Holmes E. C., Pathogen population bottlenecks and adaptive landscapes: Overcoming the barriers to disease emergence. Proc. Biol. Sci. 283, 20160727 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abraham J., Corbett K. D., Farzan M., Choe H., Harrison S. C., Structural basis for receptor recognition by New World hemorrhagic fever arenaviruses. Nat. Struct. Mol. Biol. 17, 438–444 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fernandez-Leiro R., Scheres S. H. W., A pipeline approach to single-particle processing in RELION. Acta Crystallogr D Struct. Biol. 73, 496–502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zivanov J., et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pettersen E. F., et al. , UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 59.de la Rosa-Trevín J. M., et al. , Scipion: A software framework toward integration, reproducibility and validation in 3D electron microscopy. J. Struct. Biol. 195, 93–99 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Waterhouse A., et al. , SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Afonine P. V., Headd J. J., Terwilliger T. C., Adams P. D., PHENIX news. Comput. Crystallogr. Newsl. 4, 43–44 (2013). [Google Scholar]
- 62.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goddard T. D., et al. , UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiao C., Rossmann M. G., Interpretation of electron density with stereographic roadmap projections. J. Struct. Biol. 158, 182–187 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.