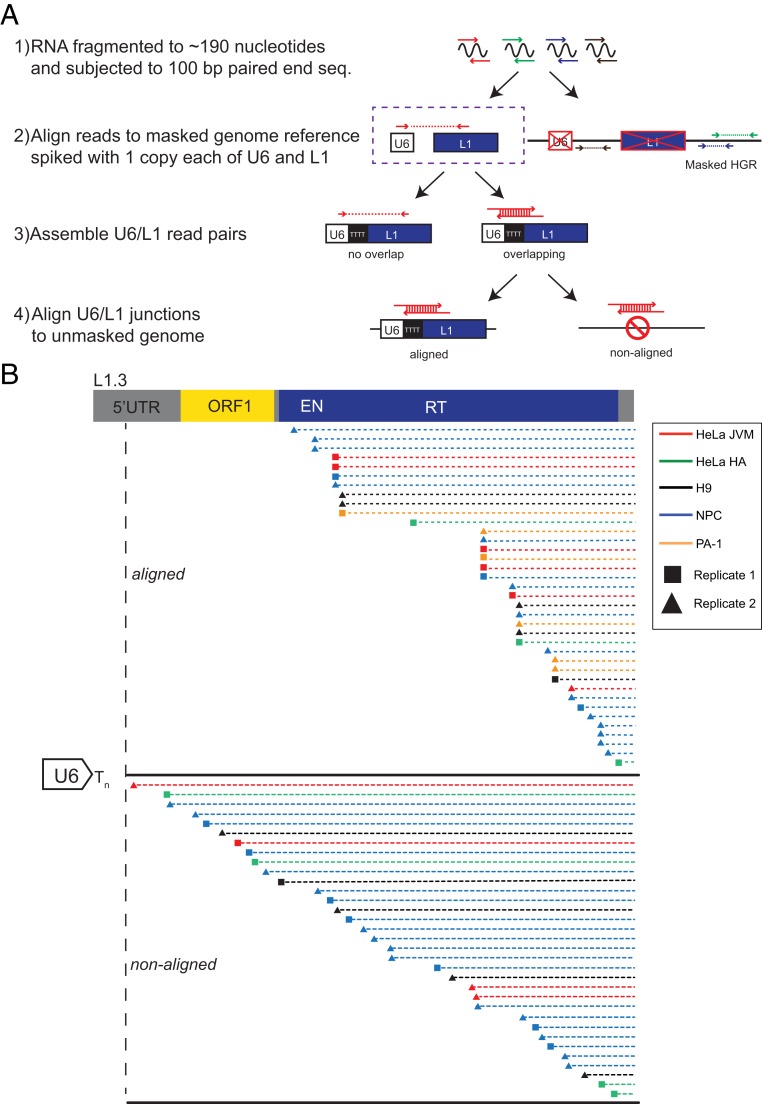
Fig. 4.
RNA-seq detection of endogenous U6/L1 chimeric RNAs in human cell lines. (A) Rationale of the RNA-seq experiments. Step 1: Ribosome-depleted RNA was fragmented to ∼190 nucleotides and subjected to 100-bp paired-end DNA sequencing. Step 2: RNA-seq read pairs (arrows) were aligned to a repeat masked version human reference genome (HGR/build Grch38), which contained “spiked-in” copies of a single U6 (white rectangle) and single L1.3 (blue rectangle) sequence. RNA-seq reads that did not map to U6 or L1 were discarded from subsequent analyses. Step 3: Overlapping U6/L1 read pairs were merged to determine the U6/L1 junction sequences. U6/L1 read pairs that contained a gap (i.e., “no overlap”) were discarded from subsequent analyses. Step 4: Overlapping U6/L1 junctions were aligned to the unmasked HGR. If the U6/L1 junction mapped to the HGR with >90% accuracy, it was designated as an “aligned” read. U6/L1 junctions that failed to map to the HGR with at least 90% accuracy were designated as “non-aligned” reads. (B) Structures of RNA-seq U6/L1 junctions. A schematic of a full-length RC-L1 is indicated at the top. The general structure of a U6/L1 chimeric junction sequence consists of the 3′ end of a U6 snRNA cDNA sequence ending in ∼4 to 8 thymidine nucleotides (left side of figure; white arrow ending in Tn) conjoined to a variably 5′−truncated L1 sequence. Two independent RNA-seq libraries were generated from HeLa-JVM, H9, NPC, and PA-1 cells, respectively (squares and triangles, respectively). One RNA-seq library was generated from HeLa-HA cells. Red horizontal lines, HeLa-JVM; green horizontal lines, HeLa-HA; yellow horizontal lines, PA-1; black horizontal lines, H9; blue horizontal lines, human NPCs. Each horizontal dashed line represents a single U6/L1 junction RNA-seq merged sequence read. The triangle or square at the left end of the horizontal dashed lines indicates the approximate location of the U6/L1 junction point relative to L1.3. The top set of dashed lines represent 16 U6/L1 junction sequences that mapped (“aligned”) to the HGR. The bottom set of dashed lines represent 33 U6/L1 junction sequences that did not map (“non-aligned”) to the HGR. These 33 U6/L1 junctions contained a copy of U6 conjoined to an L1 present in the same transcriptional orientation. The remaining 4 U6/L1 chimeras that did not map to the HGR (SI Appendix, Table S6) contained a copy of U6 conjoined to an L1 present in the opposite transcriptional orientation.