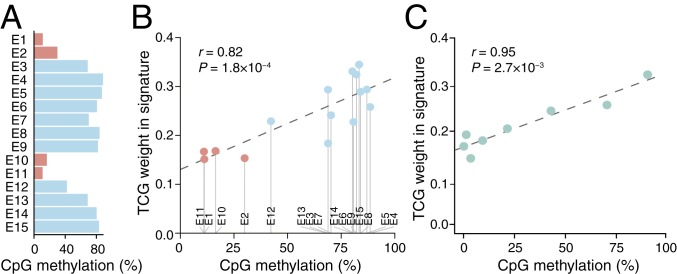
Fig. 3.
UV trinucleotide signature in promoters vary with methylation. (A) Extensive variability in CpG methylation across ChromHMM chromatin states. (B) Positive correlation between methylation level and the weight of TCG > TTG substitutions in the trinucleotide signature (local genome normalized) across chromatin states. (C) The weight of TCG > TTG substitutions in the trinucleotide signature of promoters varies positively with methylation level. Annotated coding gene promoters with sufficient bisulfite coverage (n = 12,239) were divided into 10 methylation level bins (the first three, all representing 0%, were merged into one; x axis indicates average methylation). Pearson’s correlation coefficients across regions/bins are indicated in B and C.