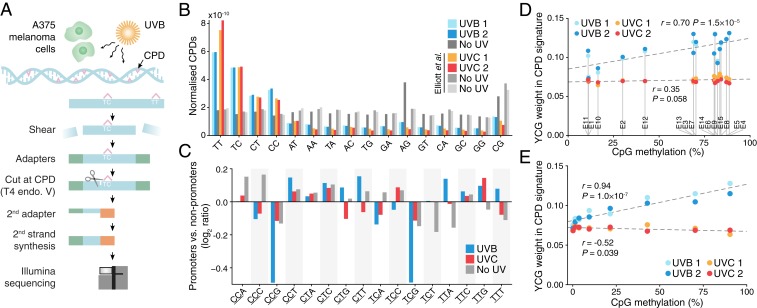
Fig. 4.
Genome-wide mapping of UVB-induced pyrimidine dimers reveals reduced DNA damage in promoters with reduced CpG methylation. (A) Simplified protocol overview for genome-wide mapping of CPDs induced by UVB in A375 human melanoma cells. Cells were treated with 10,000 J/m2 UVB (310 nm), and DNA was harvested immediately for analysis. (B) Genome-wide CPDs counts for all dinucleotides, normalized with respect to genomic dinucleotide counts and library size, showing preferable detection at dipyrimidines as expected. UVC results from Elliott et al. (15) were included for comparison. (C) Reduced UVB-induced DNA damage at YCG sites in promoters. Comparison of the CPD trinucleotide signature (relative formation frequency per genomic site) in highly expressed promoters compared to nonpromoter regions, expressed as a log2 ratio. Examined patterns include one additional 3ʹ base following the CPD-forming dipyrimidine (underscored) in all possible combinations, to enable comparison between CpG- and non-CpG–adjacent sites. Results are shown for UVB, UVC, and no-UV controls. (D) CPD frequency at YCG (weight in CPD trinucleotide signature) increases with increasing CpG methylation across ChromHMM regions, specifically for UVB. (E) CPD frequency at YCG increases with increasing CpG methylation across annotated promoters, specifically for UVB. Bins were defined as in Fig. 3C. Pearson’s correlation coefficients across regions/bins are indicated in D and E.