Figure 1.
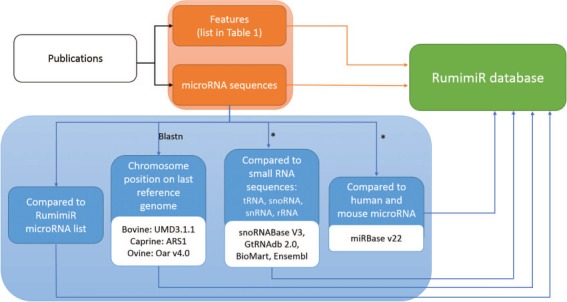
Procedure followed to include microRNAs and their associated features in the RumimiR database. From each publication (black rectangle), the microRNA sequences and their features (orange rectangles) were extracted and included in the RumimiR database. Several analyses were performed to expand the description of the microRNAs collected (blue rectangles): determination of chromosomal position in the latest reference genome of each species (using the NCBI Blastn tool), search for redundancy with microRNAs already present in the RumimiR database, search for identity with other small microRNAs to compare the sequences, search for identity with human or mouse microRNAs present in miRBase. *, using a home-made Python script (Supplementary Figure 2).
