Figure 1.
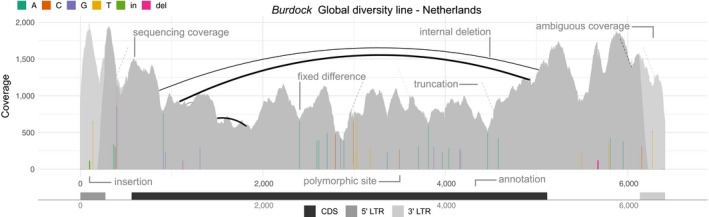
Example of the visualization of TE diversity with DeviaTE using burdock from D. melanogaster. Sequencing coverage is shown separately for unambiguously (dark grey) and ambiguously (light grey) mapped reads. Fixed differences and polymorphic sites are shown as coloured bars, with the height of the bar corresponding to the frequency of the SNP. The reference allele is not shown in the visualization. Internal deletions are displayed as arcs, where the width of the arcs scales with the abundance of the deletion. Terminal deletions are shown as dashed lines, with their opacity indicating the abundance of the deletion (darker lines indicate higher abundance). An annotation of the TE is shown at the bottom. Note that ambiguously mapped regions coincide with the long LTRs of burdock. Data are from a D. melanogaster line caught in the Netherlands (Grenier et al., 2015)
