Figure 3.
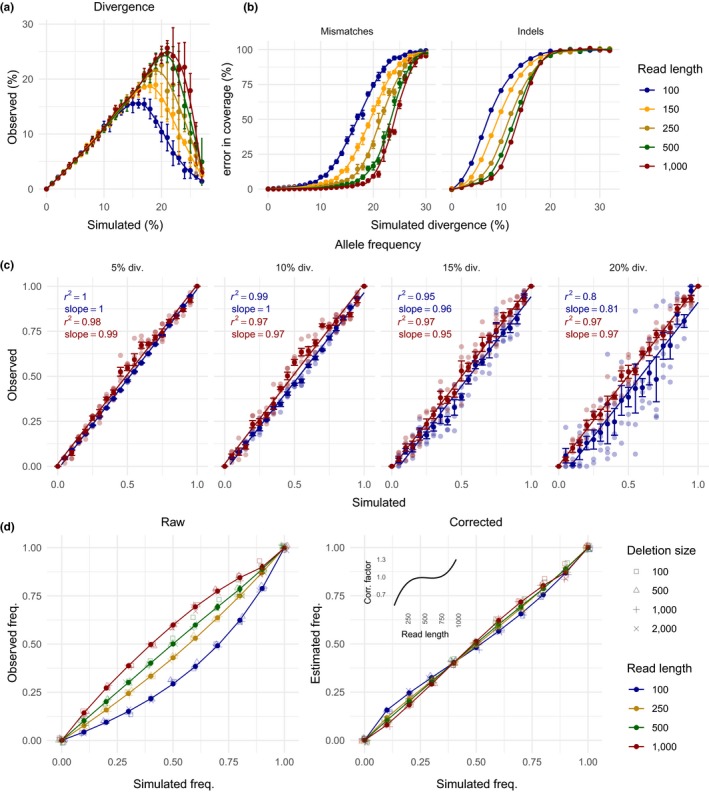
Validation of DeviaTE with simulated data. (a) Comparison between simulated and observed sequence divergence. DeviaTE accurately recovers simulated divergence of up to 15% for short reads (100 bp) and 22% for long reads (1,000 bp). Notably, the accuracy does not increase linearly with the read length. (b) Error of the estimated coverage dependent on the simulated divergence of reads. DeviaTE accurately reproduces the simulated coverage if the mismatch rate is smaller than 8% and 16% for short and long reads, respectively. Lower divergence levels are tolerated for indels. (c) Accuracy of allele frequency estimates dependent on the divergence. DeviaTE accurately reproduces allele frequencies of SNPs up to a divergence of 15%. (d) Accuracy of estimated frequencies of internal deletions. Since raw frequency estimates show a small bias (left), we implemented a read length dependent correction factor (right, inset), which substantially improves the accuracy of frequency estimates (right). Note that in a, c, and d a diagonal would indicate perfect agreement between expected and observed values
