Figure 5. Unsupervised co-editing network analysis.
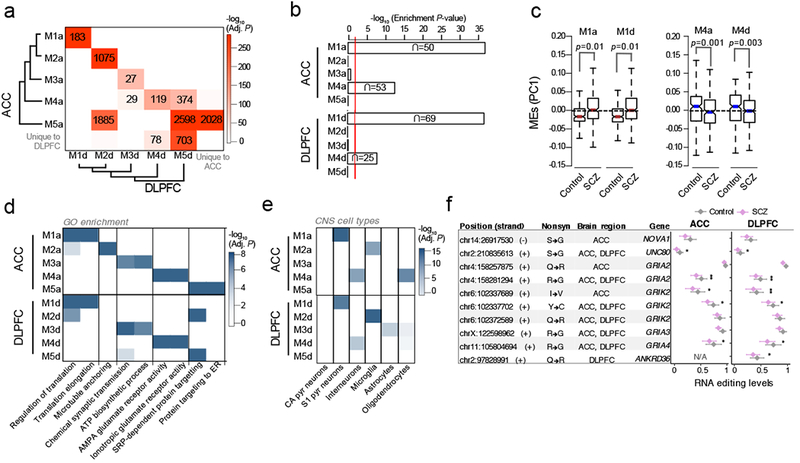
(a) Overlap analysis of co-editing modules identified within the ACC and DLPFC. Unsupervised clustering was used to group modules by module eigengene (ME) values using Pearson’s correlation coefficient and Ward’s distance method. Significance of overlap was computed (one-sided Fisher exact test, Bonferroni correction) and p-values were colored on a continuous scale (bright red, strongly significant; white, no significance). The number of overlapping sites are displayed in each cell with a significant overlap. (b) Enrichment analysis of differentially edited sites within co-editing networks (one-sided hypergeometric test). (c) Assessment of ME values for modules M1a and M1d (over-edited) and M4a and M4d (under-edited). Differential ME analysis was conducted using a linear model and covarying for age, RIN, PMI, sample site and gender. (d) The top functional enrichment terms and (e) brain cell-type enrichment results for all identified modules, verifying similar functional and cell-type properties of co-editing networks in the ACC and DLPFC. Enrichment was computed using one-sided hypergeometric test and adjusted for multiple comparisons using Bonferroni correction. (f) A collection of nonsynonymous sites within SCZ-related AMPA glutamate receptor modules M4a and M4d (** indicates Adj. P <0.05, * indicates P < 0.05 derived from differential RNA editing analysis, see Table S1 for details). Whisker dot plots show mean and whiskers represent minimum and maximum standard error of the ACC (ncontrol=245, nSCZ=225) and DLPFC (ncontrol=286, nSCZ=254).
