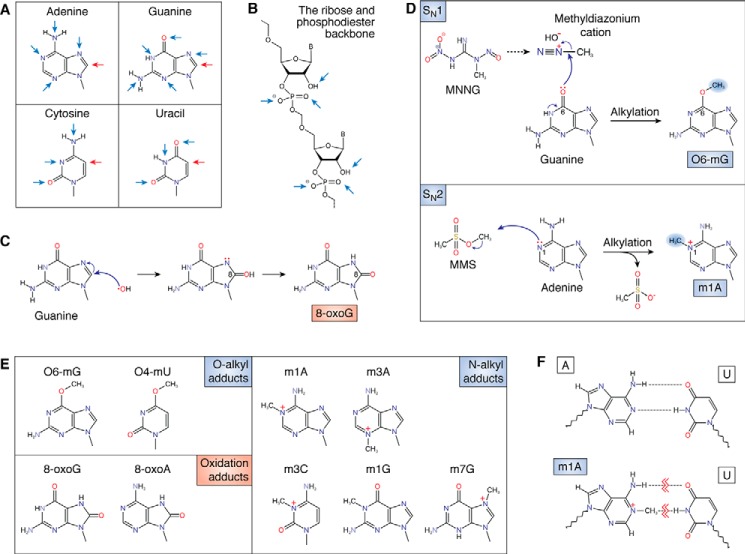
Figure 1.
Effects of alkylation and oxidation on the chemical structure of RNA. A, structures of the four RNA nucleobases with the location of common oxidation sites (red arrows) and alkylative damage sites (blue arrows) marked. B, targets of chemical insults (mainly alkylative damage) to the phosphodiester backbone and 2′-OH of the ribose. C, reaction between a hydroxyl radical and guanosine, forming the 8-oxoG adduct. D, reaction between MNNG and guanosine, which forms the O-alkyl adduct O6-mG (top). The bottom shows the reaction between methyl methanesulfonate (MMS) and adenosine forming the N-alkyl adduct m1A. E, common modified bases that result from chemical insults. Alkylative adducts, grouped by the position of modification, are as follows: O-alkyl adducts: O6-methylguanosine (O6-mG) and O4-methyluridine (O4-mU); N-alkyl adducts: N1-methyladenosine (m1A), N3-methyladenosine (m3A), N3-methylcytidine (m3C), N1-methylguanosine (m1G), and N7-methylguanosine (m7G); oxidative adducts: 8-oxo-7,8-dihydroguanosine (8-oxoG) and 8-oxo-7,8-dihydroadenosine (8-oxoA). F, damaged nucleobases disrupt base pairing. The base pairing between adenosine and uridine (top) is disrupted by the formation of the alkylative adduct m1A (bottom).