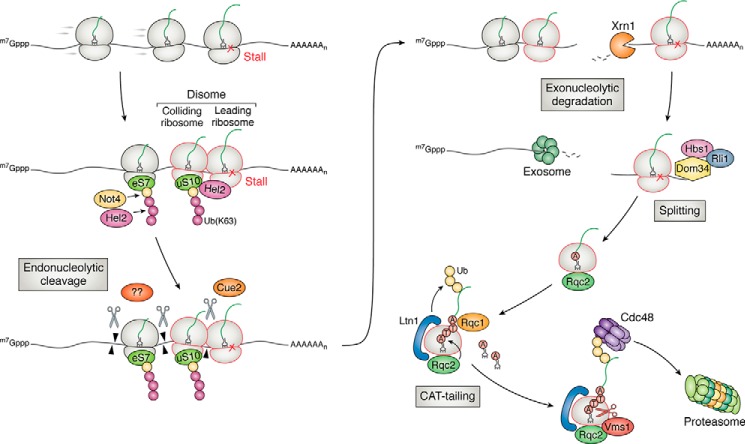
Figure 3.
Overview of ribosome-based quality control of aberrant mRNAs in yeast. Ribosomes stall on an aberrant transcript (such as a damaged one), resulting in ribosome collisions. The unique structural feature of the collided ribosomes is recognized by the E3 ligase Hel2 for ubiquitination of multiple targets, including uS10 and eS7 on the 40S subunit. The ubiquitinated ribosomes are recognized by a number of factors, which are hypothesized to recruit an unknown endonuclease to cleave the mRNA and initiate NGD. In a secondary branch of NGD, Cue2 cleaves the mRNA in the A site of the collided ribosome. The cleaved transcript is degraded by Xrn1 and the exosome. The resulting ribosomes are rescued by Dom34, Hbs1, and Rli1. The incomplete peptide attached to the peptidyl-tRNA on the 60S subunit is recognized and ubiquitinated by another E3 ligase Ltn1. C-terminal alanine and threonine residues (CAT tails) are added to the nascent peptide by Rqc2 to help expose lysine residues to the active site of Ltn1 and/or mark the nascent peptide for degradation in an Ltn1-independent manner. Released from the ribosome by Vms1, the ubiquitinated polypeptides are presented to the proteasome for degradation by Cdc48 as facilitated by Rqc1.