Abstract
Activation-induced deaminase (AID) and apolipoprotein B mRNA-editing enzyme catalytic subunit (APOBEC) enzymes convert cytosines to uracils, creating signature mutations that have been used to predict sites targeted by these enzymes. Mutation-based targeting maps are distorted by the error-prone or error-free repair of these uracils and by selection pressures. To directly map uracils created by AID/APOBEC enzymes, here we used uracil-DNA glycosylase and an alkoxyamine to covalently tag and sequence uracil-containing DNA fragments (UPD-Seq). We applied this technique to the genome of repair-defective, APOBEC3A-expressing bacterial cells and created a uracilation genome map, i.e. uracilome. The peak uracilated regions were in the 5′-ends of genes and operons mainly containing tRNA genes and a few protein-coding genes. We validated these findings through deep sequencing of pulldown regions and whole-genome sequencing of independent clones. The peaks were not correlated with high transcription rates or stable RNA:DNA hybrid formation. We defined the uracilation index (UI) as the frequency of occurrence of TT in UPD-Seq reads at different original TC dinucleotides. Genome-wide UI calculation confirmed that APOBEC3A modifies cytosines in the lagging-strand template during replication and in short hairpin loops. APOBEC3A's preference for tRNA genes was observed previously in yeast, and an analysis of human tumor sequences revealed that in tumors with a high percentage of APOBEC3 signature mutations, the frequency of tRNA gene mutations was much higher than in the rest of the genome. These results identify multiple causes underlying selection of cytosines by APOBEC3A for deamination, and demonstrate the utility of UPD-Seq.
Keywords: DNA repair, base excision repair (BER), ChIP-sequencing (ChIP-seq), mutagenesis, DNA sequencing, AID/APOBEC, cytosine deaminase, UPD-Seq, uracilome
Introduction
Uracils may be introduced in DNA in at least three different ways (1, 2). First, cellular water causes cytosines to deaminate at a low rate and it is estimated that this creates ∼80 uracils per haploid human genome per day (3). Second, bacterial and eukaryotic DNA polymerases readily utilize dUTP instead of TTP during replication (4–6). Viruses such as HIV that infect macrophages with high dUTP levels (7) accumulate up to 500 uracils per viral genome (8) and this inhibits viral integration in the host genome (8, 9).
The third pathway for the acquisition of uracils in DNA is through the action of the AID/APOBEC3 family of ssDNA-specific (ssDNA-specific) cytosine deaminases found in most vertebrates (10–13). These enzymes confer innate or acquired immunity against infections, and inhibit the spread of mobile genetic elements principally through the conversion of cytosines in DNA or RNA to uracils. In addition to their immune function, the AID/APOBEC enzymes also cause “off-target” effects in the cellular genome including base substitution mutations (14–16) and translocations (17–19). For example, APOBEC3A (A3A) and APOBEC3B (A3B) are frequently expressed at high levels in different cancer types and the genomes of these tumors carry mutations characteristic of these enzymes (16, 20–23).
Although such studies show that accumulation of uracils in viral and cellular genomes result in diverse biological outcomes, most investigations of AID/APOBEC enzymes use mutations as proxies for uracils. This is problematic because uracils in DNA are excised by the ubiquitous uracil-DNA glycosylase (Ung) and the resulting abasic sites may be repaired through error-prone or error-free pathways. This is probably why human tumors expressing one of these enzymes at very high levels show only modestly higher uracil levels than tumors with low expression levels (24, 25). The uracils may also be replicated without repair (26–28) and the presence of such U:A pairs cannot be detected using most DNA sequencing technologies. Finally, restoration of cytosines at these uracils through base-excision repair (BER) erases the evidence of the existence of uracils. Consequently, a better understanding of how AID/APOBEC enzymes target specific genomic regions will come only with direct mapping of uracils in DNA.
Although there is no established method for the mapping of uracils in DNA, Bryan et al. (29) described two closely related methods to map uracils in DNA that depend on the generation of double-strand breaks at uracils by combining Ung with endonuclease IV, and have used it to map uracils in the HIV-1 genome (30). This method requires that the DNA is highly uracilated (>1% of cytosines converted to uracil) (29, 31) and is not applicable to situations where expression of AID/APOBEC enzymes cause only small increases in genomic uracil levels (a few uracils per 106 bp) (32). A different method replaces the uracil with a biotinylated uracil or cytosine using complete BER and uses the biotin tag to pulldown the DNA fragments for sequencing (33) and requires a large number of biochemical steps (Fig. S1), which are likely to introduce artifacts. We describe below a simpler biochemical method to isolate uracilated DNA fragments that may be applied to genomes with low levels of uracils. Furthermore, using this method we demonstrate that A3A preferentially targets lagging strand templates during replication, as well as many hairpin loops and tRNA genes in the Escherichia coli genome. The targeting of tRNA genes by A3A is consistent with mutational studies in yeast expressing AID/APOBECs and analysis of whole-genome sequencing (WGS) of human tumors.
Results
A method to pulldown uracilated DNA fragments
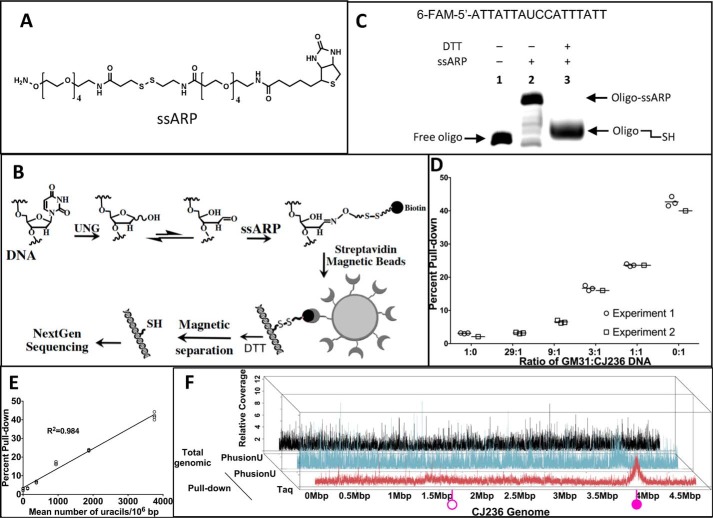
We created a new method for pulldown of uracilated DNA fragments for sequencing (uracil pulldown sequencing; UPD-seq). This technology involves the use of a disulfide link-containing chemical, ssARP (34) (Fig. 1A), to tag abasic sites created by the removal of uracils from DNA. The biotin within ssARP is bound to streptavidin beads to pulldown these tagged fragments from those lacking uracils and the reduction of the disulfide link separates the pulled down fragments from streptavidin (Fig. 1B). This methodology was tested on a fluorescently labeled DNA oligomer containing a single uracil. The products created by the binding of ssARP to DNA following excision of the uracil and subsequent reduction of the disulfide linkage by DTT were electrophoresed. The resulting gel showed that the DNA nearly quantitatively reacted with ssARP and the treatment of the bound complex with DTT separated the DNA from the biotin tag (Fig. 1C).
Figure 1.
Labeling, pulldown, and sequencing of uracil-containing DNA. A, structure of ssARP. The molecule contains a reactive amine at one end and a biotin at the other end with a disulfide linkage within the connecting linker. B, schematic presentation of ssARP-based pulldown of uracil-containing DNA. Uracil containing DNA is treated with Ung to excise uracils and create abasic sites. The abasic sites are reacted with ssARP and the resulting biotinylated DNA is bound with streptavidin-coated magnetic beads. The unbound DNA is removed using magnetic separation. DNA bound to the magnetic beads is released from the beads using DTT. The released DNA is subjected to next-generation sequencing. C, confirmation of ssARP labeling and release of DNA using DTT. The sequence of the 6-Fam-labeled DNA oligomer used is shown at the top. The uracils were excised using Ung and the DNA was reacted with ssARP (lane 2) and subsequently treated with DTT (lane 3). The reaction products were separated on a denaturing gel. D, relationship between genomic uracil levels and efficiency of DNA pulldown. The DNAs from genomes with low (GM31) and high (CJ236) levels were mixed at different ratios (x axis), the fragments containing uracils were labeled using ssARP and pulled down. The x axis shows these ratios. The percentage of DNA pulled down from different samples in two different experiments is plotted. E, the data from D were plotted as a function of the mean uracil content of each sample. F, depth of coverage of sequencing reads across the genome of CJ236. Total genomic DNA libraries amplified by PhusionU polymerase (black), UPD-seq using PhusionU (light blue), or Taq polymerase (burgundy). The positions of the origin of replication and terminus are, respectively, indicated by filled and open purple lollipops on the on the reference genome (x axis).
To test UPD-seq on uracilated genomic DNA, we chose two different E. coli strains: GM31 (WT; i.e. ung+ dut+), which contain few uracils (2 to 4 uracils/106 bp) (32) and CJ236 (ung− dut−), which accumulates a very high uracil level (∼3,600 uracils/106 bp) (32, 35). In two separate experiments, the GM31 and CJ236 DNAs were mixed at different ratios and the pulldown was performed. The results showed that higher amounts of CJ236 DNA reproducibly resulted in higher percentages of DNA being pulled down (Fig. 1D). Furthermore, there was a linear relationship between percent pulldown and of the average number of uracils in these DNA mixtures (Fig. 1E).
Uracils in E. coli CJ236 genome
To demonstrate that UPD-seq could be used to map uracils across a genome, we subjected DNA pulled down from CJ236 genome to NextGen sequencing. One concern was that the DNA released from the streptavidin beads contains a chemical scar (Fig. 1B) that is a modified form of an AP site. Different DNA polymerases are known to polymerize base insertion across AP sites, albeit at different efficiencies compared with normal bases (36–38). Hence, we compared the copying of a DNA template containing either a uracil or a chemical scar with three different polymerases, Dpo4, PhusionU, and Taq. The results showed that all three polymerases performed synthesis across both the uracil and the scar (Fig. S2). Despite this, it is reasonable to expect that the scarred DNA may be amplified less efficiently than any DNA without scars that is carried over during the pulldown. We selected PhusionU and Taq polymerases for the amplification of pulldown genomic fragments from CJ236 instead of Dpo4, because the latter enzyme is known to be error-prone (39).
All three libraries (total genome-PhusionU, and uracil pulldown with PhusionU, or Taq polymerase) covered the CJ236 genome (Fig. 1F). In particular, the uracil pulldown libraries prepared using PhusionU and Taq polymerases, respectively, covered 97 and 99% of the genome (Table S1). This is consistent with the expectation that CJ236 has an elevated level of dUTP due to a mutation in the dut gene (5) and hence dUMP is incorporated throughout the genome. Interestingly, the library prepared using the Taq polymerase (and to a lesser extent using PhusionU) had a peak around the origin of replication (Fig. 1F, filled pink lollipop, and Fig. S3). As the E. coli cells were grown in rich medium, they should contain multiple replication forks (40) and this is the likely explanation for the higher density of uracil pulldown fragments near the origin of replication compared with other parts of the genome. Together, these results show that the uracil pulldown and sequencing strategy outlined in Fig. 1B works despite the chemical scar left behind by ssARP in DNA.
UPD-seq of E. coli expressing human APOBEC3A
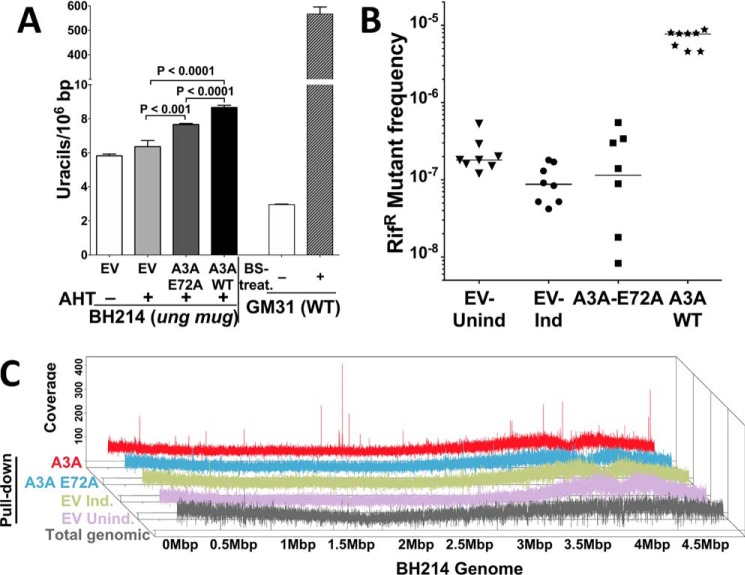
The human APOBEC3A gene was expressed in an E. coli strain defective in both its uracil-DNA glycosylases, Ung and Mug, and the level of genomic uracils and mutations were monitored. As negative controls, the empty vector (EV) plasmid and a catalytically defective mutant of A3A (E72A) were also evaluated in these experiments. This strain, BH214, had slightly higher uracil levels than its WT parent (GM31; ung+ mug+) and expression of A3A in this strain caused a modest (∼36%), but statistically significant, increase in genomic uracils (Fig. 2A). The E72A mutant of A3A also caused a small increase in genomic uracils in this case, but the resulting level was lower than WT A3A (Fig. 2A) suggesting that the mutation creates a defect in catalysis but may not completely inactivate A3A. The ability of overexpressed A3A E72A mutant to slightly increase genomic uracils was unexpected, but reproducible (Fig. S4). As expected, expression of WT A3A caused a large increase in the frequency of rifampicin-resistant mutants compared with both mutant A3A and EV (Fig. 2B) confirming the mutagenicity of A3A.
Figure 2.
Uracils and mutations caused by A3A. A, quantification of genomic uracils in the strain BH214. E. coli genomic DNA from an ung+ strain (GM31) with or without bisulfite treatment, respectively, serve as positive and negative controls. AHT, inducer anhydrotetracycline; BS, bisulfite. B, rifampicin-resistant mutant frequency for BH214 cells containing empty vector, plasmid containing gene for WT A3A or A3A mutant E72A. Mutant frequencies from eight independent cultures are shown with the median value is indicated by a horizontal line. Cells with empty vector were either uninduced (EV-UnInd) or induced (EV-Ind). C, depth of coverage of sequencing reads from different libraries across the genome of BH214. Uracil pulldown libraries: A3A–induced (red), catalytically dead mutant A3A-E72A–induced (light blue), empty vector-induced (olive), empty vector-uninduced (lavender), and libraries from total genomic DNA (gray).
Uracilated DNA fragments from these strains were subjected to UPD-seq (Fig. 1B) and the reads were aligned to the BH214 sequence. In all cases, there was coverage of the entire E. coli genome (Fig. 2C) suggesting that either some uracils were being accumulated throughout the genome in this strain, which is defective in uracil excision, or that there was some carry-over of DNA without uracils in the pulldown. Regardless, the sequence profile of the pulldown from cells expressing WT A3A was substantially different from all other profiles and showed several peaks (Fig. 2C, top line, Fig. S5).
Analysis of genome-wide uracilation at TC dinucleotides
We created a simple measure of the frequency of A3A conversion of any cytosine in the genome to uracil by A3A called the uracilation index (UI). This is the fraction of the sequencing reads at a cytosine that are called as thymine multiplied by 1,000 (Fig. S6). This measure counts uracils that were not excised by Ung and will be read as thymines by the Taq polymerase, and uracils that were converted to a chemical scar during the pulldown. The Taq polymerase overwhelmingly inserts an adenine across abasic sites (41, 42) effectively replacing the original cytosine with a thymine. When the average UI was calculated at all TC sequences in the BH214 genome for different sequencing runs, it was lowest for WGS of the strain with EV plasmid (1.06) and somewhat higher for UPD-seq of cells with the EV plasmid or A3A mutant plasmid (1.24 and 1.21, respectively). The higher UI of UPD-seq is likely to be due to the enrichment of uracil-containing DNA during the pulldown. The average UI was highest for UPD-seq of BH214 with the WT A3A plasmid (1.45) and reflects increased conversion of cytosines by A3A to uracils (Fig. 2A).
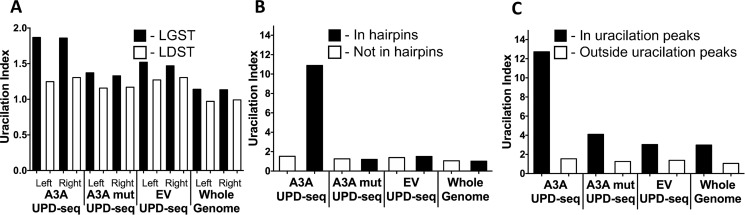
Although the increase in average UI at TCs due to A3A was small, it showed a replicative strand bias that was seen previously for C to T mutations caused by APOBECs in E. coli (A3G) (43), yeast (A3A and A3B) (44), and cancer genomes (45, 46). In both the left and the right replichores of the E. coli genome of cells expressing WT A3A, the UI of TCs in the lagging-strand template (LGST) was 1.4 to 1.5 times the UI for the leading-strand template (LDST; Fig. 3A). The UPD-seq of genomes from A3A mutant- or EV-containing cells, and the whole genome sequence of BH214 with EV also showed a strand bias, but the magnitude of this bias was smaller (1.1 to 1.2; Fig. 3A). The uracilation seen in cells lacking an active A3A is likely due to water-mediated cytosine deaminations, and has been described previously to cause a replicative strand bias in C to T mutations (43). These results show that the UI index is useful for detecting DNA strand preferences of A3A, without a need to isolate and sequence individual mutants.
Figure 3.
Uracilation index at TC dinucleotides calculated for three sets of UPD-seq (A3A, A3A-E72A, and EV) and one WGS (EV) data. The index is plotted for (A) the left and right replichores and separated for targeted cytosines in the LDST or LGST. B, TC dinucleotides in predicted hairpins and those are predicted not to lie in hairpins. C, TC dinucleotides inside and outside the uracilation peaks.
A recent publication (47) showed that A3A prefers to deaminate cytosines in hairpin loops and A3A signature passenger mutations in cancer genomes are highly correlated with TCs in predicted strong hairpins with small loops. Specifically, the cancer mutation study defined a mathematical term called expected relative mutation frequency (fexp) for different potential hairpin loops, and found that hairpin loops with fexp ≥ 4x above the nonhairpin baseline were likely to acquire mutations that were attributable to A3A (47). These results suggested that UI should be higher for TCs in loops of such predicted hairpins in the E. coli genome. When the BH214 genome was analyzed using the same criteria, 2,185 TCs were found to have fexp ≥ 4x (Table S2). The UI of TCs within these hairpins was seven times higher in UPD-seq of A3A expressing cells compared with UI of TCs in the rest of the genome (Fig. 3B). In contrast, the UPD-seq of cells with A3A mutant or EV plasmid, or WGS of EV plasmid cells showed little difference between TCs in predicted hairpins and the rest of the genome (Fig. 3B). This shows that the presence of active A3A in cells increases the conversion of TCs in hairpin loops to TUs.
Identification of peaks within UPD-seq data
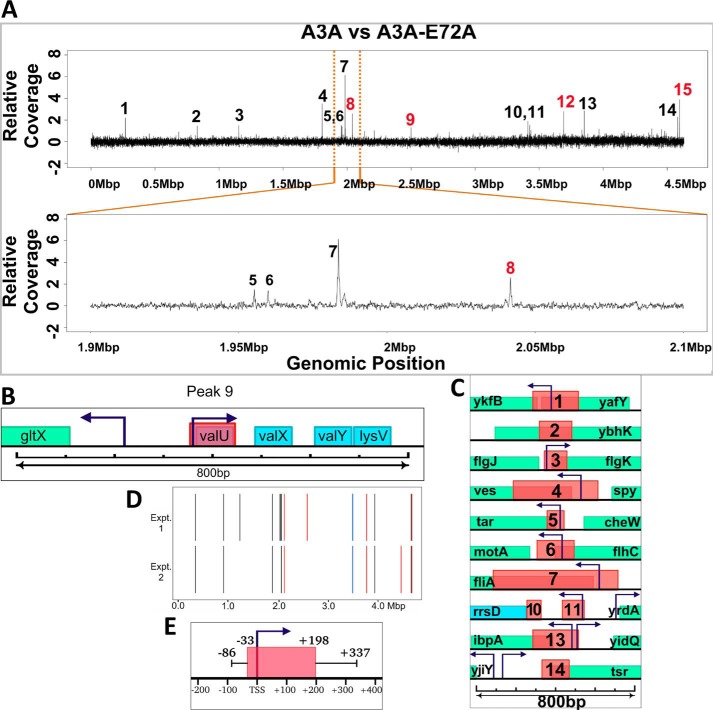
As noted above, the plots of the depth of coverage in UPD-seq data against the genomic position showed a presence of peaks for WT A3A that were absent in the other two pulldown libraries (Fig. 2C). To eliminate any variations in the pulldown efficiencies or ease of amplification due to local sequence variations, the UPD-seq data were normalized using two different algorithms (NDC and MACS) (48). The software compared the UPD-seq data from cells expressing WT A3A with the two negative controls and identified uracilated peaks caused by A3A. The two methods gave a very similar distribution of peaks (Fig. S7) and the two sets of peaks overlapped (Fig. S8). The intersection of results using NDC and MACS was used to define uracilated peaks, and this identified 15 peaks (Fig. 4A). The UI of these 15 regions together was much higher than the UI of the rest of the genome in all the genomic libraries (Fig. 3C). UI for the peaks was about eight times higher than the rest of the genome for WT A3A expressing cells, whereas this ratio was between two and three for other libraries (Fig. 3C). This suggests that the regions represented by the peaks accumulated uracils created by A3A or water much more frequently than other genomic regions and this is why they are over-represented in the pulldown libraries.
Figure 4.
Uracilation peaks created by A3A. A, normalized differential coverage plot of A3A-induced versus A3A-E72A–induced libraries. The peaks are numbered and the peaks that overlap tRNA genes are in red. At the bottom, a region within the E. coli genome between 1.9 and 2.1 Mb has been expanded to provide a clearer view of peaks 5 to 8. B, the genes in and near peak 9. The salmon-colored box indicates the width of the uracil-containing peak. Green represents a protein-coding gene and blue represents RNA-coding genes. The start and direction of transcription are indicated by arrows. The size scale appears below the genes. C, all the peaks that overlap with protein-coding genes are shown. The salmon-colored boxes indicate the widths of the uracil-containing peaks. Green represents protein-coding genes and blue represents RNA-coding genes. D, comparison of genomic positions of peaks in two different experiments. Peaks overlapping with tRNA, rRNA, and protein-coding genes are, respectively, shown in red, blue, and black. E, median distances of upstream and downstream edges of peaks relative to TSS are shown. The whiskers show the first quartile upstream of start and third quartile downstream of end of peaks.
Genomic features of the pulldown peaks
Twenty-two genes overlapped the 15 peaks and there was a very strong bias within this group in favor of tRNA genes. The E. coli genome contains about 4,300 protein-coding genes (CDS; 4,035,915 bp), 22 rRNA genes (96,603 bp), and 87 tRNA genes (20,436 bp) (49), but UPD-seq preferentially enriched parts of only 12 protein-coding genes and one rRNA gene (Table S3). In contrast, six tRNA genes were found within the peaks and two of the peaks (peaks 8 and 15) contained both CDSs and tRNA genes. Hence, the apparent pulldown of the CDSs within these two peaks may be due to the adjacent tRNA genes.
Another feature of these peaks was that nearly all of them overlap the 5′-ends of transcription units. For example, peak 9 overlaps with an operon that contains four tRNA genes of which three code identical valine tRNAs (Fig. 4B) (49). Despite this, the peak contains only the 5′-end valU gene (Fig. 4B, salmon-colored box). A similar strong bias favoring 5′-ends of transcription units was also seen within the CDSs and the rRNA gene found within the peaks. All except one of these peaks (peak 2) occurs near the 5′-end of a gene and most contain transcription start site (TSS) of one or two genes (Fig. 4C).
Reproducibility of UPD-seq identified peaks
To confirm the reproducibility of this technique, all the steps of UPD-seq, starting with cell growth and extraction of genomic DNA, were repeated using the same plasmids in the same strain (BH214) and related plasmids in a different strain, BH212. The results from the second experiment using BH214 were analyzed using the criteria described above, and this showed 12 uracilated peaks. Eleven of these peaks overlapped with peaks identified in the first experiment (Fig. 4D; p value 1.3 × 10−9) and 16 genes were common in the two sets (Table S3). This shows that uracilated peaks identified using UPD-seq are highly reproducible.
Although the strain BH214 lacks both uracil-excising glycosylases, Ung and Mug, BH212 is ung− mug+ (50). Consequently, the Mug glycosylase may eliminate some of the uracils and hence some uracilated peaks created by A3A. This prediction was confirmed when UPD-seq of BH212 cells expressing A3A showed fewer uracilated peaks (10) and genes (14) compared with BH214 (Fig. S9). Despite this, uracilated peaks in BH212 contained a total of seven tRNA genes confirming the strong bias of A3A toward tRNA genes (Table S4). Six of the peaks seen in BH212 genome overlapped with BH214 peaks, and the two pulldowns shared six peaks and six genes (Fig. S9 and Table S4). Together, these results show that some regions in the E. coli genome are intrinsically good targets of A3A.
Confirmation of A3A targeting through genomic DNA sequencing
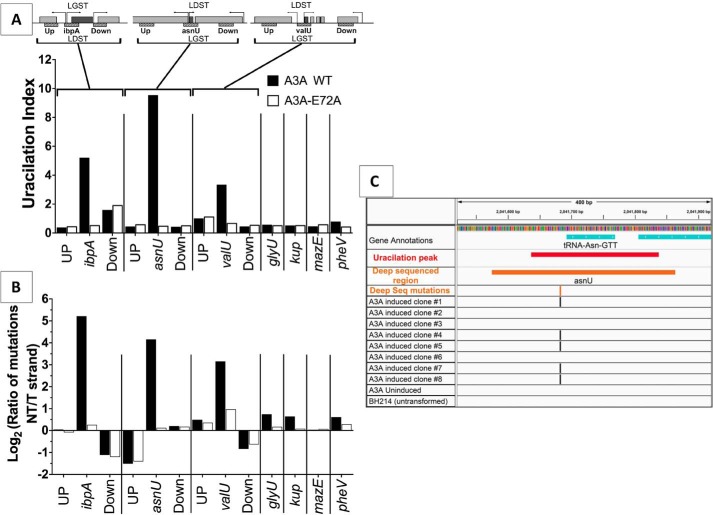
Ultra-deep sequencing is a strategy designed to detect rare mutations in a population of organisms (51) and we employed it to detect C to U conversions in the regions covered by the uracilation peaks. About 300-bp regions overlapping three of the uracilated peaks (peaks 8, 9, and 13; Fig. 4A) were amplified from genomic DNA (i.e. which had not been subjected to ssARP-based pulldown) from cells expressing A3A and the PCR products were sequenced to a mean depth of coverage of greater than 500,000. The cytosine deaminations caused by A3A were identified within these reads as C to T or G to A changes within 5′-TC/GA dinucleotides and the frequency of their occurrence was calculated. To eliminate PCR-based biases regarding amplification efficiencies or errors, we compared these results with those from genomic DNA of cells expressing the A3A mutant. As negative controls, we amplified regions upstream and downstream of each of these genes and sequenced them. Furthermore, four additional genes that were not identified in the pulldown and hence were not expected to contain excess uracils were also amplified and sequenced. The results of this analysis are summarized in Fig. 5.
Figure 5.
C:G to T:A changes within uracilation peaks. The top of the figure shows arrangements of the three genes with uracilation peaks (dark gray) and the regions upstream and downstream (light gray). The direction of transcription of each gene is shown by an arrow and replication-based distinction of the two strands within the targeted genes is indicated by the labels LGST and LDST. The genomic regions are arranged so that transcription of the gene overlapping a uracilation peak is from left to right. The ∼300-bp regions amplified and sequenced are shown by hatched boxes and are labeled Up, name of gene with uracilation peak, or Down. The genes in these amplified regions are: ibpA, yidQ (Up) and ibpB (Down); for asnU, yeeO (Up) and Cbl (Down); valU, gltX (Up) and xapR (Down). The valU operon also contains valX, valY, and lysV genes downstream of valU. A, the fraction of C:G to T:A changes in the 5′-TC context normalized to the number of TC sequences in each region. B, the log2 of the ratio of C to T changes in the NT to template strand (T) for different genes. C, visualization of the 400-bp sequence around the asnU gene. Successive lines show the span of the uracilation peak (red), the deep sequenced region (orange), and the position of the C to T changes in uracilation (orange) and mutations in whole genome sequence of eight independent clones expressing A3A (black), one clone with no induction of A3A and one clone without A3A.
All three regions identified by the pulldown showed C:G to T:A changes with the greatest degree of conversions seen in the asnU gene (∼1% of 5′-TC/GA converted to TT/AA) (Fig. 5A). Furthermore, the frequency of conversions was 5 to 20 times lower in DNA from cells expressing the inactive A3A mutant showing that the changes were caused by the ability of A3A to convert cytosines to uracils. In contrast, the upstream and downstream regions of these three A3A-targeted genes showed much lower frequency of C to T conversions and there was little difference between the frequency in WT versus mutant expressing cells (Fig. 5A). This confirms that the uracilation peaks at these three regions were confined to a narrow region near the 5′-ends of these genes. The four other genes subjected to deep sequencing, glyU, kup, mazE, and pheV, did not appear as peaks in UPD-seq and showed much lower frequency of C:G to T:A changes in deep sequencing with little difference in frequencies for DNAs from cells expressing WT or mutant A3A (Fig. 5A). Together, these results confirm that the original genomic DNA contained a much higher frequency of A3A-dependent C to U conversions in the very regions enriched in the pulldown compared with the neighboring regions or genes not enriched in the pulldown. Furthermore, the uracils in these genomic regions were created by the catalytic activity of A3A.
In all three A3A-targeted regions, the C to T changes overwhelmingly occurred in the nontemplate strand of the gene (Fig. 5B). It is known that cytosines in the nontemplate (NT) strand are more accessible to water-mediated deamination during transcription (52, 53) and hence this suggests that transcription may allow access to the NT strand for A3A. It should be noted that this strand bias did not always match with the previously described replication-based strand bias. Although the NT strand in the ibpA gene is also LGST, the NT strand for asnU and valU genes is the LDST (Fig. 5, top panel). Therefore, transcription may play a much bigger role than replication in the creation of strand bias in the genes that contain uracilation peaks created by A3A.
Analysis of mutations in independent clones expressing A3A
The peaks identified in Fig. 4 cover only about 0.01% of the E. coli genome and hence very few mutations created by A3A should be found in these peaks, if they were randomly distributed across the genome. On the other hand, if A3A selectively creates uracils in the uracilation peak regions, then this should result in an increase in C:G to T:A mutations within these regions. To test this, a culture of BH214 cells containing A3A plasmid was split into eight cultures that were then induced for A3A expression. After overnight growth, the cells were plated and one colony was picked from each of the eight cultures. Additionally, one colony from a noninduced culture and one from cells without the A3A plasmid were also picked. All 10 colonies were subjected to WGS.
Among all the mutations in the eight A3A expressing cultures, nearly 80% were C:G to T:A transitions and of these 11% were within one of the 15 uracilation peaks (Table 1). Thus, the A3A in these clones showed about a 1,000-fold preference for causing mutations in the regions identified by UPD-seq (Table 1). For example, five of eight clones acquired a C to T mutation at the same position upstream of the asnU gene (Fig. 5C) and a substantial fraction of the independent clones also acquired mutations at a single cytosine within ibpA (4 of 8) and valU genes (2 of 8; Fig. S10). Remarkably, many of the deep sequencing reads of these regions also contained C to T changes at these same positions. Specifically, 32, 11, and 9% of the deep sequencing reads, respectively, for the asnU, ibpA, and valU genes contained thymine at the cytosines that were frequently mutated in the independent clones (Fig. 5C and Fig. S10). Together, these results show that the uracilation peaks contain specific cytosines that are targeted by A3A at high frequencies causing C:G to T:A mutations.
Table 1.
Distribution of C:G to T:A mutations
Cumulative mutations in the eight independent clones expressing A3A.
| Number of bp | Number of mutations |
p valueb | ||
|---|---|---|---|---|
| Expecteda | Observed | |||
| In uracilation peaks | 4,398 | 0.44 | 54 | 2.2 × 10−13 |
| Outside uracilation peaks | 4,626,741 | 478 | 424 | |
a Normalized to number of C:G pairs in the sequences.
b Chi-squared test with Yates' continuity correction.
Lack of correlation between high transcription and uracilated peaks
As most of the uracilation peaks contained 5′-ends of genes and the deep sequencing showed a bias in mutations that is most easily explained by transcription, we considered the possibility that A3A may be targeting highly transcribed genes. When all the E. coli genes were put into 10 bins based on the level of transcription, most genes that were pulled down using ssARP were in the lowest transcription bin (Fig. S11A). Dividing E. coli genes into quartiles based on transcription level also found only about one-third of the genes in the highest quartile (Fig. S11B). We also compared the transcription levels of the tRNA genes enriched in the BH212 and BH214 pulldowns with the levels for nonenriched tRNA genes and found the former group had a slightly higher level of transcription, but the difference was not statistically significant (Fig. S11C). Thus, despite the presence of TSS and 5′-ends of genes within the peaks identified by UPD-seq and the strand bias in mutations within the peaks identified by deep sequencing, there is a lack of strong correlation between high transcription levels of genes and their presence in the peaks.
Uracil-enriched genes are not correlated with RNA:DNA hybrids
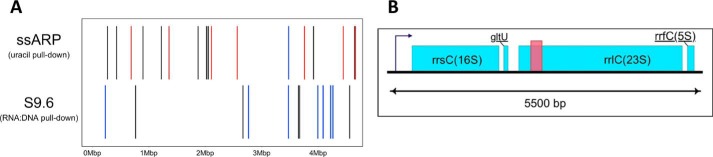
It is well-established that AID targets immunoglobulin switch regions that contain R-loops (54, 55) and it has been suggested that the APOBEC enzymes may also access ssDNA within R-loops (56, 57). To identify RNA:DNA hybrid regions in the E. coli genome, the BH214 DNA fragments were pulled down using the mAb against RNA-DNA hybrids, S9.6 (58), and sequenced. When these sequences were mapped to the E. coli genome they did not overlap with any of the peaks of uracilated regions in either BH212 or BH214 (Fig. 6A and Table S5). The antibody did not pulldown any of the tRNA genes, but instead pulled down fragments from at least three 23S rRNA genes (Tables S5 and S6).
Figure 6.
Peaks from the pulldown of RNA:DNA hybrids using S9.6 antibody. A, genomic positions of peaks from ssARP (uracil) and S9.6 (RNA:DNA) pulldown experiments. Peaks overlapping with tRNA, rRNA, and protein-coding genes are, respectively, shown in red, blue, and black. B, example of a peak from the S9.6 pulldown. The salmon-colored box indicates the width of the uracil-containing peak. The rrsC, rrlC, and rrfC are rRNA genes and gltU is a tRNA gene.
Our results are consistent with reports about yeast (59, 60) and human cells (61), which also found that rRNA genes are prone to the formation of RNA:DNA hybrids. An additional feature of the peaks in the S9.6 pulldown was that these peaks frequently did not include the 5′-ends of the rRNA operon transcripts or the tRNA genes contained within these operons. In the example shown, the uracilated peak (Fig. 6B, salmon-colored box) lies in the middle of the rRNA operon and does not overlap a tRNA gene that lies between the 16S and 23S RNA genes. This lack of overlap between the regions pulled down using ssARP and S9.6, and the differences in the types of genic and intergenic regions in the two pulldowns suggest that A3A does not target genomic regions containing RNA-DNA hybrids.
Discussion
Salient features of targeting of genomic regions by A3A
We have described here a biochemical method, UPD-seq, to map uracils in genomic DNA that is similar in principle to methods for mapping chromosomal transcription factor-binding sites (e.g. ChIP-seq) (62) or epigenetic marks (MeDIP-Seq) (63). However, in contrast to these other methods that typically use an antibody, we use an enzyme and a chemical to replace the uracils with a cleavable biotin tag. The method was used to identify biological processes, DNA structures, genes, and genomic regions where A3A preferentially creates uracils. Analysis of these pulldown sequences confirmed a number of different patterns of targeting by A3A.
The pulldown sequences from cells expressing catalytically active A3A contained more frequent occurrences of C to T changes within TC sequences than the pulldowns from A3A mutant or EV controls. This was quantified as uracilation index (UI) and used to confirm that A3A preferentially converts cytosines in the LGST to uracils compared with LDST (Fig. 3A). This suggests that the ssDNA in LGST is accessible to A3A and this is consistent with mutational results in E. coli, yeast, and cancer genomes (43–46).
The UI was also used to show that A3A targets TCs in hairpin loops that were predicted to be good targets based on analysis of cancer genome mutations. Overall, the UI of the TCs in these loops was seven times the UI of TCs in the rest of the genome. When UIs of all TCs in hairpin loops were analyzed for the stability of the stem of the hairpin, UI increased with the strength of the stem (Fig. S12A). Among these hairpins, loops 3 nt in size had the highest UI and this preference was more pronounced for loops with high stem strength (>12) (Fig. S12B). Furthermore, the cytosines were more susceptible to deamination if they were present in the 3′-half of the loops, especially at high stem strength (Fig. S12C). These patterns are consistent with the biochemistry of A3A and the occurrence of A3A signature mutations in hairpin loops in cancer genomes (47).
We used two computational algorithms, two negative controls, and a very strong selective criterion, a signal that is >5σ above background noise, to identify regions that were over-represented in the pulldown (Fig. 4A). The results obtained from two different runs for one E. coli strain and using a different bacterial strain identified many of the same genes (Fig. 4D and Figs. S7 and S9). This demonstrates the reproducibility of the UPD-seq technology and shows that it identifies genomic regions that are highly susceptible to acquiring uracils through cytosine deaminations by A3A.
The important features of these uracilated regions include a large overrepresentation of tRNA genes (Tables S3 and S4) and the presence of 5′-ends of transcription units of most genes within these regions (Fig. 4, B and C). Most of the uracilated peaks included TSS and extended 33 bp upstream and 198 bp downstream (median values of upstream and downstream edges of peaks relative to TSS; Fig. 4E). Furthermore, deep sequencing of the original genomic DNA for three of the regions identified by UPD-seq confirmed that they had acquired excess C:G to T:A mutations (Fig. 5A) and showed that the mutated cytosines were predominantly in the NT strand (Fig. 5B). Although these data suggest a role for transcription in the A3A targeting, other data (Fig. S11) show that high-level transcription is not necessary for targeting. Our data are also inconsistent with the formation of RNA:DNA loops as being the cause of A3A targeting (Fig. 6). Together, these results show that the 5′-ends of a small number of E. coli genes, among which tRNA genes are greatly over-represented, are highly preferred by A3A for cytosine deamination.
Although these genomic regions were highly susceptible to deamination, they do not share any obvious structural features. When the nucleic acid-folding software Mfold (64) was used to predict likely secondary structure of the 15 uracilation peak regions, a wide variety of structures were predicted (Fig. S13). Furthermore, the cytosines that were most frequently found as thymines in UPD-seq of A3A expressing cells were often in predicted stems, instead of loops (e.g. peaks 1 and 2, Fig. S13). In some peaks, the frequently deaminated cytosines were in loops much larger than 6 nt (e.g. peaks 3 and 6, Fig. S13) and hence these regions are quite unlike any that have been described as preferential targets for A3A in cancer genome mutation studies (47). It will be important to elucidate the reasons for selective targeting of these regions by A3A in future studies.
AID/APOBECs cause mutations in tRNA genes in yeast
Our results are largely consistent with mutational studies of AID/APOBEC enzymes in yeast (57, 65–67). Yeast expressing human APOBEC1, AID, A3A, A3B, or A3G (21, 57, 66) or a lamprey homolog of AID/APOBEC, PmCDA1 (57, 65), showed elevated frequencies of mutations in many genes. Although there were some differences between the results of these studies, they found that the frequency of mutations was highest in promoters, near TSS and 5′-untranslated regions of genes, and it fell off rapidly in the CDS and 3′-untranslated regions of the genes (21, 57, 66). These studies also found a strong (66) or modest (57, 67) preference for mutations at cytosines in the nontemplate strand compared with the template strand. These features are quite similar to those of uracilation peaks created by A3A in E. coli.
Furthermore, two of the yeast studies found highly preferential targeting of genes transcribed by RNA polymerase III (pol III) including tRNA genes, compared with the RNA polymerase II (pol II)-transcribed genes (66, 67). The mutations caused by these enzymes were often clustered and the frequency of mutations in tRNA and other pol III-transcribed genes was much higher than the protein-coding genes, especially in the case of hyperactive AID (67) and A3B (66). In particular, Saini et al. (66) estimated that the rate of mutation in a tryptophan tRNA gene was 70 times higher than in the pol II-transcribed canavanine-resistance gene. Together with UPD-seq experiments described above these studies suggest that many of the AID/APOBEC family enzymes have an affinity for structures at the 5′-ends of actively transcribed tRNA genes including their promoters. Whether the deaminases bind these regions because they are in a persistent single-stranded state or form some specific structures is currently unknown.
APOBEC signature mutations in tRNA genes in human tumors
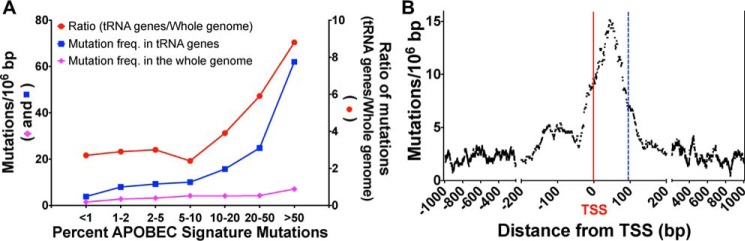
These mutational results in yeast and our uracil mapping results in E. coli suggested that human tumors that overexpress A3A or A3B (A3A/B) may also suffer mutations within tRNA genes. To confirm this, we examined WGS from three datasets: 2279 whole-genomes from the Pan-cancer analysis of whole genome (PCAWG) dataset (68), a set of 560 breast cancer whole genomes (69), and a set of 126 whole genomes from the Broad Institute (70). To distinguish between mutations caused by A3A/B and those from other sources; the tumors were divided into different classes based on the percentage of their total mutations that were of the APOBEC signature (14, 15, 71). Both the frequency of mutations in the tRNA genes and in the whole genome increased as the percentage of mutations with APOBEC signature increased, but the increase was much larger for the tRNA genes (Fig. 7A). As the percentage of APOBEC signature mutations increased from ≤10 to >50%, the ratio of the mutation frequency in tRNA genes to the mutation frequency in the whole genome increased from about 3 to nearly 9 (Fig. 7A). The mutations in the tRNA genes were focused narrowly within the genes, beginning a few bp before the TSS and extending just beyond the 3′ edge of the tRNA genes (Fig. 7B). The distribution of mutations in these tumors is remarkably similar to the distribution of A3A-generated uracils identified in the pulldown of E. coli genomic fragments (Figs. 4, B and E) suggesting that the same molecular mechanism drives both distribution patterns. A more complete analysis of tRNA gene mutations in human cancers will be presented elsewhere.
Figure 7.
Frequency and distribution of tRNA gene mutations in human tumors. A, the tumors in the PCAWG database were divided into different classes based on the frequency of mutations with the APOBEC signature. The frequency of all the mutations across the whole genome and in the tRNA genes are shown for each class (left y axis). The ratio of frequencies of tRNA gene mutations to all the mutations is also shown (right y axis). B, the frequency of mutations in and around tRNA genes is shown, combining data from 472 APOBEC-positive tumors from the PCAWG, Broad, and Sanger databases. The TSS is marked with a red vertical line and is numbered “0.” The TSS for most tRNA genes lies a few nucleotides upstream of the 5′ end of the mature tRNA and most tRNA genes are <75 bp long. A few tRNA genes are longer by up to 20 bp because of the variable loop and the intronic sequences. The position of the 3′-end of the longest tRNA gene is shown by a dashed blue vertical line. Note that the x axis is divided into three segments.
In summary, we have developed and validated a method that uses one enzyme and one chemical to define granular details within a very modestly increased level of uracils in the E. coli genome due to A3A. Unlike mutational studies, it does not require isolation of mutant clones and is ideally suited for mapping uracils created by the AID/APOBEC deaminases. The preference of A3A for LGST and hairpins with short loops found here is consistent with experimental work in yeast and analysis of cancer genome mutations, and this shows that A3A behaves in E. coli in a manner very similar to what it does in mammalian cells. These results help restrict molecular models that can explain how A3A finds cytosine substrates, and has found a group of regions in the genome that is highly selected by A3A for unknown reasons. One of the predictions from our study was that A3A should target tRNA genes in the human genome. We confirmed this prediction through analysis of tumor genomes demonstrating that UPD-seq methodology is simple, but powerful, and has predictive capabilities.
Experimental procedures
Bacterial strains and plasmids
E. coli K12 strains CJ236 (F′ chloramphenol-resistant; ung− dut−), GM31 (dcm6 thr1 hisG4 leuB6 rpsL ara14 supE44 lacY1 tonA31 tsx78 galK2 galE2 xyl5 thi1 mtl1), BH214 (GM31 ung− mug−), and BH212 (GM31 ung− (λUP81)) (50) were used in this study. Human APOBEC3A gene and its E72A mutant (72) were cloned into pASK-IBA5C (IBA Lifesciences) and pGEX-6P2 (GE Healthcare) plasmid vectors. The resulting clones were validated using Sanger sequencing (DNA sequencing core, University of Michigan).
Labeling an oligonucleotide with ssARP
A 17-mer oligo (17 UCC-56FAM-ATTATTAUCCATTTATT; IDT) was treated with E. coli Ung for 30 min at 37 °C. The resulting abasic sites were labeled with EZ-Link Alkoxyamine-PEG4-SS-PEG4-Biotin (ssARP; Thermo Scientific) at pH 7.4 for 1 h at 37 °C. To break the disulfide linkage, the samples were treated with 100 mm DTT for 5 min at 37 °C. Finally, an equal volume of formamide dye was added and the samples were heated at 95 °C for 5 min and chilled on ice. Labeling reactions were analyzed on a 20% denaturing polyacrylamide gel.
Lesion bypass assay
Two primer/template hybrids were constructed using the oligomers: 5′-CCA GCT CGG TAC CGG GCT UGC CTT TGG AGT CGA CCT GCA GAA TCC GCC GCG-3′ and 5′-56FAM-CGC GGC GGA TTC TGC AGG TCG ACT CCA AAG G-3′ (IDT). To construct a uracil-containing primer/template hybrid, the oligomers were hybridized together with the first molecule in 2-fold molar excess. The same primer/template hybrid was also sequentially treated with Ung (1 h at 37 °C), ssARP (1 h at 37 °C), and DTT (10 min at 37 °C) to create a hybrid with a chemical scar where uracil was originally present. The primer was extended using Dpo4 (a kind gift from Dr. L. J. Romano, Wayne State University), Taq polymerase (Promega), or PhusionU polymerase (ThermoFisher Scientific). The reaction mixtures were heated to 95 °C and cooled to 40 °C. Then dNTPs were added to the reaction mixtures and incubated at 40 °C for 1 min. Formamide containing dye was added to quench the reaction. The products were electrophoresed in a 20% denaturing PAGE gel.
Quantification of genomic uracils
The quantification of uracils in DNA was done using AA6 (32, 73). Briefly, genomic DNA was digested with HaeIII and treated with O-allylhydroxylamine (10 mm; Sigma) to block the endogenous abasic sites. It was sequentially treated with uracil-DNA glycosylase and AA6 (2 mm). Click reaction was performed by adding DBCO-Cy5 (1.6 μm, Sigma), and the DNA was spotted on a nylon membrane. The membrane was scanned with Typhoon FLA 9500 and the quantification of fluorescence intensities was performed using ImageQuant software.
Cell growth and expression of A3A
The BH214 cells containing pASK plasmids were grown overnight in medium containing chloramphenicol. The overnight cultures were diluted 100-fold in growth media and incubated in a shaker at 37 °C for 2 h. Following their 10-fold dilution in the selective growth media, A3A expression was induced using 0.5 μg/ml of anhydrotetracycline (Cayman Chemical), and grown for about 4 h until the cell cultures reached A600 of 0.9.
The BH212 cells containing the pGEX plasmids were grown overnight in media containing carbenicillin. The overnight cultures were diluted ×100 in fresh media and were grown until A600 reached 0.5. The cultures were diluted again in fresh media and A3A expression was induced by adding isopropyl 1-thio-β-d-galactopyranoside to 1 mm (Gold Biotechnology) and the cultures were grown until the A600 reached 0.5.
Rifampicin-resistance mutational assay
LB medium with chloramphenicol was inoculated with independent colonies of BH214 cells containing different plasmids and the cells were grown overnight. Each culture was diluted 100-fold in the growth media and incubated in a shaker at 37 °C for 2 h. This was followed by an 8-fold dilution of each culture and the expression of A3A was induced using 0.5 μg/ml of anhydrotetracycline (Cayman Chemical). In the case of BH214 with empty vector, each culture was split into two following overnight growth and AHT was added to only one of the two cultures. All cultures were grown until the cultures reached A600 of ∼0.8. Different dilutions of cells were spread on LB plates with 100 μg/ml of rifampicin (Sigma) or without the antibiotic. The plates were incubated overnight at 37 °C and colonies were counted. Rifampicin-resistant mutant frequencies were calculated as the ratio of the number of cfu/ml on rifampicin-containing plates divided by the number of cfu/ml on LB plates.
Pulldown of uracil-containing genomic DNA
During the technique development stage of this work, E. coli genomic DNA was digested with HaeIII restriction endonuclease enzyme, and the enzyme was removed using phenol:chloroform extraction and ethanol precipitation. Pre-existing abasic sites in DNA were blocked by treating with 10 mm methoxyamine (Sigma) for 1 h at 37 °C and then incubated with Ung for 1 h at 37 °C to excise the uracils and create new abasic sites. The DNA was incubated with 10 mm ssARP for 1 h at 37 °C and purified by phenol:chloroform (1:1) extraction and ethanol precipitation. This biotinylated DNA was pulled-down using Dynabeads MyOne Streptavidin C1 magnetic beads (Invitrogen). The beads were separated from the solution on a magnetic stand (DynaMag, Invitrogen), and the supernatant containing the unbound DNA was removed. Magnetic beads were washed twice with the binding and wash buffer provided by the suppliers of the beads, and the beads were resuspended in 1× TE buffer. The bound DNA was released by incubating the beads with 100 mm DTT for 10 min at 37 °C. The beads were placed on the magnetic stand, and the DNA in the free solution was concentrated using ethanol precipitation. The DNA pellet was dissolved in 0.1× TE buffer, and its concentration was determined by NanoDrop 2000c (Thermo Scientific).
Enrichment of RNA/DNA hybrids from genomic DNA
The enrichment of RNA/DNA hybrid containing DNA fragments was performed as described previously (74). Briefly, genomic DNA was fragmented with HaeIII restriction enzyme treatment and the enzyme was removed by phenol:chloroform extraction and ethanol precipitation. The digested DNA (10 μg) was incubated with S9.6 antibody (Kerafast; 5 μg) in 400 μl of IP buffer (10 mm sodium phosphate, pH 7.0, 140 mm NaCl, 0.1% Tween 20) at 4 °C for 2 h. The Dynabeads protein G (ThermoFisher Scientific) (10 μl) was added to the reaction and incubated again for 2 h at 4 °C. The beads were magnetically separated and washed three times with the IP buffer. The beads were resuspended in IP buffer and treated with Proteinase K (Promega) overnight at 4 °C. The immunoprecipitated DNA was purified by phenol:chloroform extraction and ethanol precipitation.
Library preparation and sequencing of pulldown DNA
The genomic DNA was sonicated to produce 500-bp fragments (Covaris S2). DNA labeling using ssARP, and pulldown were performed as described before. DNA libraries were prepared using a TruSeq nano-DNA library preparation kit (Illumina). In some experiments, two different DNA polymerases (Taq, Promega; PhusionU, Thermo-Fisher) were used during the library amplification step. The prepared DNA libraries were sequenced on Illumina MiSeq, HiSeq (Michigan State University), or Illumina MiniSeq (Wayne State University). All the raw sequencing reads are deposited to NCBI Sequence Read Archive and can be accessed under BioProject ID PRJNA448166.
Sequence alignment and analysis
All bioinformatic analysis was performed using LINUX-based software available at the High-performance Computing Services, Wayne State University. Sequencing reads that were mapped to the plasmids (pGEX-6P-2 and pASK-IBA5C) were removed and the remaining sequencing reads were aligned to the corresponding E. coli reference sequence using BWA (version 0.7.12) (75). The plasmid sequences were removed because they contained some genomic sequences such as the lac operon genes, and the high copy number of plasmids in cells created artificial peaks at these loci in the genome.
In one of the sequencing experiments involving BH212 samples, we multiplexed our samples with libraries prepared from PCR products of genes, malE, sodC, and kan. We detected that a small percentage of sequencing reads from these PCR libraries were misassigned to other libraries causing higher coverage at these genes and were detected as peaks. These spurious peaks were eliminated by removing reads with lower quality as suggested by Wright and Vetsigian (76). Thus, reads for a Phred quality score of 28 or higher were used in this analysis. This largely eliminated the problem of contamination from the PCR products. Samtools (version 1.2-83) (75) was used to sort the aligned reads and extract genomic depth of coverage.
The uracilated genomic regions were identified using two different algorithms. MACS (version 2.1.0.20150420) (48) was used to call peaks where there was an enrichment of sequencing reads from the A3A expressing cells. Separately, a function of moving average (mav) window of 100 bp was applied to the depth of coverage and normalized differential coverage (NDC) of two sets of libraries were calculated using the following formula.
| (Eq. 1) |
R (version 3.4.1) (77) statistical package was used to calculate NDC and make the NDC and barcode plots (https://github.com/rayanramin/NDC).4 The signal threshold was set at 5σ (5 times the standard deviation of NDC) to detect peaks. The peaks detected by MACS and NDC were correlated using R package GenometriCorr (78). The p value of relative distance correlation in pairwise comparisons of identified peaks detected by MACS and NDC was calculated using the Kolmogorov-Smirnov test. The genes overlapping the peaks were identified by performing BLAST alignment of the sequences of the peaks with MG1655 sequence and the transcription start sites for the genes were obtained from the EcoCyc database (49).
UI for any set of TC dinucleotides in the genome is defined by the equation,
| (Eq. 2) |
where the summation is performed over all the TCs in the set. The set may be defined by different criteria such as TCs in a genomic region or TCs in hairpin loops of specific stem strength or loop size. Integrated Genome Viewer was used for data visualization in Fig. 5C and Fig. S10 (79).
Deep sequencing analysis
The primers used to amplify the A3A-targeted regions are listed in Table S7. Each genomic region was separately amplified, the PCR products were combined and used to prepare Illumina DNA libraries. The libraries were subjected to paired-end sequencing using an Illumina MiSeq sequencer. The sequencing reads were mapped to sequences of the genomic regions and the UI of each amplified region was calculated using the formula described above.
The strand bias in cytosine deaminations were calculated using the following formula.
| (Eq. 3) |
Prediction of hairpin loops in the E. coli genome
The BH214 genome was scanned for potential hairpin-forming sequences using the previously published procedure (47). Briefly, the genome was scanned for sequences of the form S-L-S′, where the sequences S and S′ are reverse-complementary with a sequence L intervening between them. Sequences S and S′ can bp to form a “stem,” with sequence L serving as the “loop” of the resulting stem-loop, or “hairpin” structure. For each position p in the genome, we searched for flanking sequences S and S′ such that position p would be located in the intervening loop sequence L. We required L to have a length between 3 and 11 nt. In cases where multiple alternative pairings were possible, the stem with the strongest pairing was chosen, using the shortest loop size as a tie-breaker. In this way, each position p in the genome was annotated with the following parameters: stemlen = length of the stem sequence S; looplen = length of the loop sequence L; looppos = position within L that this genomic position p occupies.
We adhered to the convention of always referring to the genomic strand in which position p is a C residue, taking the reverse complement in cases when p is a G on the genomic reference strand. We also defined a parameter stem strength, which combines information about the length and GC content of the stem sequence S, and is calculated as three times the number of G:C pairs, plus one times the number of A:T pairs in the sequence: stem strength = 3 × GC + 1 × AT.
Finally, each position in the E. coli genome was assigned a value of fexp, the expected relative mutation frequency given that position's hairpin characteristics. To calculate this value, we used the quantitative model that was determined from the genomic study of human tumor data (47), for each combination of looplen, looppos, and stem strength, we calculated the number of such positions in the human genome, and the number of observed mutations in the A3A+ tumor dataset, providing a denominator and numerator for calculating a mutation frequency that was then normalized to the overall frequency, yielding an expected relative mutation frequency.
Author contributions
A. S. B. conceptualization, data analysis, funding acquisition and project administration; R. S. and V. S. experimental work and data analysis; J. S. and M. L. W. P. experimental work; R. P.-R. data analysis; M. S. L. conceptualization and data analysis.
Supplementary Material
Acknowledgments
This work was initiated by Dr. Anjali Patwardhan in the Bhagwat lab and we thank her and other members of the lab for their contributions to the development of this project. We also thank Dr. Jared Schrader (Wayne State University) for use of his Illumina Sequencer.
This work was supported by National Institutes of Health Grant R01 GM 57200 and Wayne State University. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1–S7 and Figs. S1–S13.
The DNA sequences reported in this paper has been submitted to the NCBI Sequence Read Archive under BioProject ID PRJNA448166.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- AID
- activation-induced deaminase
- APOBEC
- apolipoprotein B mRNA-editing catalytic polypeptide-like
- ssDNA
- single-stranded DNA
- Ung
- uracil-DNA glycosylase
- UPD-seq
- uracil pulldown sequencing
- BER
- base excision repair
- NDC
- normalized differential coverage
- MACS
- model-based analysis for ChIP-Seq
- CDS
- coding sequences
- TSS
- transcription start site
- LGST
- lagging-strand template
- LDST
- leading-strand template
- NT
- nontemplate
- UI
- uracilation index
- nt
- nucleotide
- AHT
- anhydrotetracycline
- EV
- empty vector
- WGS
- whole-genome sequencing
- IP
- immunoprecipitation
- PCAWG
- Pan-cancer analysis of whole genome
- pol II
- polymerase II.
References
- 1. Sousa M. M., Krokan H. E., and Slupphaug G. (2007) DNA-uracil and human pathology. Mol. Aspects Med. 28, 276–306 10.1016/j.mam.2007.04.006 [DOI] [PubMed] [Google Scholar]
- 2. Visnes T., Doseth B., Pettersen H. S., Hagen L., Sousa M. M., Akbari M., Otterlei M., Kavli B., Slupphaug G., and Krokan H. E. (2009) Uracil in DNA and its processing by different DNA glycosylases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 563–568 10.1098/rstb.2008.0186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lindahl T., and Nyberg B. (1974) Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry 13, 3405–3410 10.1021/bi00713a035 [DOI] [PubMed] [Google Scholar]
- 4. Konrad E. B., and Lehman I. R. (1975) Novel mutants of Escherichia coli that accumulate very small DNA replicative intermediates. Proc. Natl. Acad. Sci. U.S.A. 72, 2150–2154 10.1073/pnas.72.6.2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tye B. K., Nyman P. O., Lehman I. R., Hochhauser S., and Weiss B. (1977) Transient accumulation of Okazaki fragments as a result of uracil incorporation into nascent DNA. Proc. Natl. Acad. Sci. U.S.A. 74, 154–157 10.1073/pnas.74.1.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goulian M., Bleile B., and Tseng B. Y. (1980) Methotrexate-induced misincorporation of uracil into DNA. Proc. Natl. Acad. Sci. U.S.A. 77, 1956–1960 10.1073/pnas.77.4.1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kennedy E. M., Daddacha W., Slater R., Gavegnano C., Fromentin E., Schinazi R. F., and Kim B. (2011) Abundant non-canonical dUTP found in primary human macrophages drives its frequent incorporation by HIV-1 reverse transcriptase. J. Biol. Chem. 286, 25047–25055 10.1074/jbc.M111.234047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yan N., O'Day E., Wheeler L. A., Engelman A., and Lieberman J. (2011) HIV DNA is heavily uracilated, which protects it from autointegration. Proc. Natl. Acad. Sci. U.S.A. 108, 9244–9249 10.1073/pnas.1102943108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weil A. F., Ghosh D., Zhou Y., Seiple L., McMahon M. A., Spivak A. M., Siliciano R. F., and Stivers J. T. (2013) Uracil DNA glycosylase initiates degradation of HIV-1 cDNA containing misincorporated dUTP and prevents viral integration. Proc. Natl. Acad. Sci. U.S.A. 110, E448–E457 10.1073/pnas.1219702110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Conticello S. G., Langlois M. A., Yang Z., and Neuberger M. S. (2007) DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv. Immunol. 94, 37–73 10.1016/S0065-2776(06)94002-4 [DOI] [PubMed] [Google Scholar]
- 11. Salter J. D., and Smith H. C. (2018) Modeling the embrace of a mutator: APOBEC selection of nucleic acid ligands. Trends Biochem. Sci. 43, 606–622 10.1016/j.tibs.2018.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Siriwardena S. U., Chen K., and Bhagwat A. S. (2016) Functions and malfunctions of mammalian DNA-cytosine deaminases. Chem. Rev. 116, 12688–12710 10.1021/acs.chemrev.6b00296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Swanton C., McGranahan N., Starrett G. J., and Harris R. S. (2015) APOBEC enzymes: mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov. 5, 704–712 10.1158/2159-8290.CD-15-0344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alexandrov L. B., Nik-Zainal S., Wedge D. C., Aparicio S. A., Behjati S., Biankin A. V., Bignell G. R., Bolli N., Borg A., Børresen-Dale A. L., Boyault S., Burkhardt B., Butler A. P., Caldas C., Davies H. R., et al. (2013) Signatures of mutational processes in human cancer. Nature 500, 415–421 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roberts S. A., Lawrence M. S., Klimczak L. J., Grimm S. A., Fargo D., Stojanov P., Kiezun A., Kryukov G. V., Carter S. L., Saksena G., Harris S., Shah R. R., Resnick M. A., Getz G., and Gordenin D. A. (2013) An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 45, 970–976 10.1038/ng.2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nik-Zainal S., Alexandrov L. B., Wedge D. C., Van Loo P., Greenman C. D., Raine K., Jones D., Hinton J., Marshall J., Stebbings L. A., Menzies A., Martin S., Leung K., Chen L., Leroy C., et al. (2012) Mutational processes molding the genomes of 21 breast cancers. Cell 149, 979–993 10.1016/j.cell.2012.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chiarle R., Zhang Y., Frock R. L., Lewis S. M., Molinie B., Ho Y. J., Myers D. R., Choi V. W., Compagno M., Malkin D. J., Neuberg D., Monti S., Giallourakis C. C., Gostissa M., and Alt F. W. (2011) Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell 147, 107–119 10.1016/j.cell.2011.07.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Klein I. A., Resch W., Jankovic M., Oliveira T., Yamane A., Nakahashi H., Di Virgilio M., Bothmer A., Nussenzweig A., Robbiani D. F., Casellas R., and Nussenzweig M. C. (2011) Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell 147, 95–106 10.1016/j.cell.2011.07.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gazumyan A., Bothmer A., Klein I. A., Nussenzweig M. C., and McBride K. M. (2012) Activation-induced cytidine deaminase in antibody diversification and chromosome translocation. Adv. Cancer Res. 113, 167–190 10.1016/B978-0-12-394280-7.00005-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leonard B., Hart S. N., Burns M. B., Carpenter M. A., Temiz N. A., Rathore A., Vogel R. I., Nikas J. B., Law E. K., Brown W. L., Li Y., Zhang Y., Maurer M. J., Oberg A. L., Cunningham J. M., et al. (2013) APOBEC3B upregulation and genomic mutation patterns in serous ovarian carcinoma. Cancer Res. 73, 7222–7231 10.1158/0008-5472.CAN-13-1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Taylor B. J., Nik-Zainal S., Wu Y. L., Stebbings L. A., Raine K., Campbell P. J., Rada C., Stratton M. R., and Neuberger M. S. (2013) DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. Elife 2, e00534 10.7554/eLife.00534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chan K., Roberts S. A., Klimczak L. J., Sterling J. F., Saini N., Malc E. P., Kim J., Kwiatkowski D. J., Fargo D. C., Mieczkowski P. A., Getz G., and Gordenin D. A. (2015) An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat. Genet. 47, 1067–1072 10.1038/ng.3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Henderson S., Chakravarthy A., Su X., Boshoff C., and Fenton T. R. (2014) APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumor development. Cell Rep. 7, 1833–1841 10.1016/j.celrep.2014.05.012 [DOI] [PubMed] [Google Scholar]
- 24. Burns M. B., Lackey L., Carpenter M. A., Rathore A., Land A. M., Leonard B., Refsland E. W., Kotandeniya D., Tretyakova N., Nikas J. B., Yee D., Temiz N. A., Donohue D. E., McDougle R. M., Brown W. L., et al. (2013) APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 494, 366–370 10.1038/nature11881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pettersen H. S., Galashevskaya A., Doseth B., Sousa M. M., Sarno A., Visnes T., Aas P. A., Liabakk N. B., Slupphaug G., Saetrom P., Kavli B., and Krokan H. E. (2015) AID expression in B-cell lymphomas causes accumulation of genomic uracil and a distinct AID mutational signature. DNA Repair (Amst) 25, 60–71 [DOI] [PubMed] [Google Scholar]
- 26. Burns M. B., Leonard B., and Harris R. S. (2015) APOBEC3B: pathological consequences of an innate immune DNA mutator. Biomed. J. 38, 102–110 10.4103/2319-4170.148904 [DOI] [PubMed] [Google Scholar]
- 27. Krokan H. E., Saetrom P., Aas P. A., Pettersen H. S., Kavli B., and Slupphaug G. (2014) Error-free versus mutagenic processing of genomic uracil–relevance to cancer. DNA Repair (Amst) 19, 38–47 [DOI] [PubMed] [Google Scholar]
- 28. Zanotti K. J., and Gearhart P. J. (2016) Antibody diversification caused by disrupted mismatch repair and promiscuous DNA polymerases. DNA Repair (Amst) 38, 110–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bryan D. S., Ransom M., Adane B., York K., and Hesselberth J. R. (2014) High resolution mapping of modified DNA nucleobases using excision repair enzymes. Genome Res. 24, 1534–1542 10.1101/gr.174052.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hansen E. C., Ransom M., Hesselberth J. R., Hosmane N. N., Capoferri A. A., Bruner K. M., Pollack R. A., Zhang H., Drummond M. B., Siliciano J. M., Siliciano R., and Stivers J. T. (2016) Diverse fates of uracilated HIV-1 DNA during infection of myeloid lineage cells. Elife 5, e18447 10.7554/eLife.18447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ransom M., Bryan D. S., and Hesselberth J. R. (2018) High-resolution mapping of modified DNA nucleobases using excision repair enzymes. Methods Mol. Biol. 1672, 63–76 10.1007/978-1-4939-7306-4_6 [DOI] [PubMed] [Google Scholar]
- 32. Siriwardena S. U., Perera M. L. W., Senevirathne V., Stewart J., and Bhagwat A. S. (2019) A tumor-promoting phorbol ester causes a large increase in APOBEC3A expression and a moderate increase in APOBEC3B expression in a normal human keratinocyte cell line without increasing genomic uracils. Mol. Cell. Biol. 39, e00238–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shu X., Liu M., Lu Z., Zhu C., Meng H., Huang S., Zhang X., and Yi C. (2018) Genome-wide mapping reveals that deoxyuridine is enriched in the human centromeric DNA. Nat. Chem. Biol. 14, 680–687 10.1038/s41589-018-0065-9 [DOI] [PubMed] [Google Scholar]
- 34. Song C. X., Clark T. A., Lu X. Y., Kislyuk A., Dai Q., Turner S. W., He C., and Korlach J. (2011) Sensitive and specific single-molecule sequencing of 5-hydroxymethylcytosine. Nat. Methods 9, 75–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lari S. U., Chen C. Y., Vertessy B. G., Morre J., and Bennett S. E. (2006) Quantitative determination of uracil residues in Escherichia coli DNA: contribution of ung, dug, and dut genes to uracil avoidance. DNA Repair (Amst) 5, 1407–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Choi J. Y., Lim S., Kim E. J., Jo A., and Guengerich F. P. (2010) Translesion synthesis across abasic lesions by human B-family and Y-family DNA polymerases α, δ, η, ι, κ, and REV1. J. Mol. Biol. 404, 34–44 10.1016/j.jmb.2010.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nair D. T., Johnson R. E., Prakash L., Prakash S., and Aggarwal A. K. (2011) DNA synthesis across an abasic lesion by yeast REV1 DNA polymerase. J. Mol. Biol. 406, 18–28 10.1016/j.jmb.2010.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Weerasooriya S., Jasti V. P., and Basu A. K. (2014) Replicative bypass of abasic site in Escherichia coli and human cells: similarities and differences. PLoS ONE 9, e107915 10.1371/journal.pone.0107915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boudsocq F., Iwai S., Hanaoka F., and Woodgate R. (2001) Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): an archaeal DinB-like DNA polymerase with lesion-bypass properties akin to eukaryotic poleta. Nucleic Acids Res. 29, 4607–4616 10.1093/nar/29.22.4607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cooper S., and Helmstetter C. E. (1968) Chromosome replication and the division cycle of Escherichia coli B/r. J. Mol. Biol. 31, 519–540 10.1016/0022-2836(68)90425-7 [DOI] [PubMed] [Google Scholar]
- 41. Blatter N., Prokup A., Deiters A., and Marx A. (2014) Modulating the pKa of a tyrosine in KlenTaq DNA polymerase that is crucial for abasic site bypass by in vivo incorporation of a non-canonical amino acid. Chembiochem 15, 1735–1737 10.1002/cbic.201400051 [DOI] [PubMed] [Google Scholar]
- 42. Sikorsky J. A., Primerano D. A., Fenger T. W., and Denvir J. (2007) DNA damage reduces Taq DNA polymerase fidelity and PCR amplification efficiency. Biochem. Biophys. Res. Commun. 355, 431–437 10.1016/j.bbrc.2007.01.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bhagwat A. S., Hao W., Townes J. P., Lee H., Tang H., and Foster P. L. (2016) Strand-biased cytosine deamination at the replication fork causes cytosine to thymine mutations in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 113, 2176–2181 10.1073/pnas.1522325113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hoopes J. I., Cortez L. M., Mertz T. M., Malc E. P., Mieczkowski P. A., and Roberts S. A. (2016) APOBEC3A and APOBEC3B preferentially deaminate the lagging strand template during DNA replication. Cell Rep. 14, 1273–1282 10.1016/j.celrep.2016.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Haradhvala N. J., Polak P., Stojanov P., Covington K. R., Shinbrot E., Hess J. M., Rheinbay E., Kim J., Maruvka Y. E., Braunstein L. Z., Kamburov A., Hanawalt P. C., Wheeler D. A., Koren A., Lawrence M. S., and Getz G. (2016) Mutational strand asymmetries in cancer genomes reveal mechanisms of DNA damage and repair. Cell 164, 538–549 10.1016/j.cell.2015.12.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Seplyarskiy V. B., Soldatov R. A., Popadin K. Y., Antonarakis S. E., Bazykin G. A., and Nikolaev S. I. (2016) APOBEC-induced mutations in human cancers are strongly enriched on the lagging DNA strand during replication. Genome Res. 26, 174–182 10.1101/gr.197046.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Buisson R., Langenbucher A., Bowen D., Kwan E. E., Benes C. H., Zou L., and Lawrence M. S. (2019) Passenger hotspot mutations in cancer driven by APOBEC3A and mesoscale genomic features. Science 364, eaaw2872 10.1126/science.aaw2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., and Liu X. S. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Keseler I. M., Mackie A., Santos-Zavaleta A., Billington R., Bonavides-Martínez C., Caspi R., Fulcher C., Gama-Castro S., Kothari A., Krummenacker M., Latendresse M., Muñiz-Rascado L., Ong Q., Paley S., Peralta-Gil M., et al. (2017) The EcoCyc database: reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 45, D543–D550 10.1093/nar/gkw1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Beletskii A., and Bhagwat A. S. (2001) Transcription-induced cytosine-to-thymine mutations are not dependent on sequence context of the target cytosine. J. Bacteriol. 183, 6491–6493 10.1128/JB.183.21.6491-6493.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Flaherty P., Natsoulis G., Muralidharan O., Winters M., Buenrostro J., Bell J., Brown S., Holodniy M., Zhang N., and Ji H. P. (2012) Ultrasensitive detection of rare mutations using next-generation targeted resequencing. Nucleic Acids Res. 40, e2 10.1093/nar/gkr861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Beletskii A., and Bhagwat A. S. (1996) Transcription-induced mutations: increase in C to T mutations in the nontranscribed strand during transcription in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 93, 13919–13924 10.1073/pnas.93.24.13919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jinks-Robertson S., and Bhagwat A. S. (2014) Transcription-associated mutagenesis. Annu. Rev. Genet. 48, 341–359 10.1146/annurev-genet-120213-092015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yu K., Chedin F., Hsieh C. L., Wilson T. E., and Lieber M. R. (2003) R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nature Immunol. 4, 442–451 10.1038/ni919 [DOI] [PubMed] [Google Scholar]
- 55. Yu K., Roy D., Bayramyan M., Haworth I. S., and Lieber M. R. (2005) Fine-structure analysis of activation-induced deaminase accessibility to class switch region R-loops. Mol. Cell. Biol. 25, 1730–1736 10.1128/MCB.25.5.1730-1736.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Adolph M. B., Love R. P., Feng Y., and Chelico L. (2017) Enzyme cycling contributes to efficient induction of genome mutagenesis by the cytidine deaminase APOBEC3B. Nucleic Acids Res. 45, 11925–11940 10.1093/nar/gkx832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lada A. G., Kliver S. F., Dhar A., Polev D. E., Masharsky A. E., Rogozin I. B., and Pavlov Y. I. (2015) Disruption of transcriptional coactivator Sub1 leads to genome-wide re-distribution of clustered mutations induced by APOBEC in active yeast genes. PLoS Genet. 11, e1005217 10.1371/journal.pgen.1005217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Boguslawski S. J., Smith D. E., Michalak M. A., Mickelson K. E., Yehle C. O., Patterson W. L., and Carrico R. J. (1986) Characterization of monoclonal antibody to DNA·RNA and its application to immunodetection of hybrids. J. Immunol. Methods 89, 123–130 10.1016/0022-1759(86)90040-2 [DOI] [PubMed] [Google Scholar]
- 59. El Hage A., French S. L., Beyer A. L., and Tollervey D. (2010) Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 24, 1546–1558 10.1101/gad.573310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wahba L., Costantino L., Tan F. J., Zimmer A., and Koshland D. (2016) S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 30, 1327–1338 10.1101/gad.280834.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ginno P. A., Lott P. L., Christensen H. C., Korf I., and Chédin F. (2012) R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell 45, 814–825 10.1016/j.molcel.2012.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mundade R., Ozer H. G., Wei H., Prabhu L., and Lu T. (2014) Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond. Cell Cycle 13, 2847–2852 10.4161/15384101.2014.949201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li N., Ye M., Li Y., Yan Z., Butcher L. M., Sun J., Han X., Chen Q., Zhang X., and Wang J. (2010) Whole genome DNA methylation analysis based on high throughput sequencing technology. Methods 52, 203–212 10.1016/j.ymeth.2010.04.009 [DOI] [PubMed] [Google Scholar]
- 64. Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lada A. G., Stepchenkova E. I., Waisertreiger I. S., Noskov V. N., Dhar A., Eudy J. D., Boissy R. J., Hirano M., Rogozin I. B., and Pavlov Y. I. (2013) Genome-wide mutation avalanches induced in diploid yeast cells by a base analog or an APOBEC deaminase. PLoS Genet. 9, e1003736 10.1371/journal.pgen.1003736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Saini N., Roberts S. A., Sterling J. F., Malc E. P., Mieczkowski P. A., and Gordenin D. A. (2017) APOBEC3B cytidine deaminase targets the non-transcribed strand of tRNA genes in yeast. DNA Repair (Amst) 53, 4–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Taylor B. J., Wu Y. L., and Rada C. (2014) Active RNAP pre-initiation sites are highly mutated by cytidine deaminases in yeast, with AID targeting small RNA genes. Elife 3, e03553 10.7554/eLife.03553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cancer Genome Atlas Research, Network, Weinstein J. N., Collisson E. A., Mills G. B., Shaw K. R., Ozenberger B. A., Ellrott K., Shmulevich I., Sander C., and Stuart J. M. (2013) The cancer genome atlas pan-cancer analysis project. Nat. Genet. 45, 1113–1120 10.1038/ng.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nik-Zainal S., Davies H., Staaf J., Ramakrishna M., Glodzik D., Zou X., Martincorena I., Alexandrov L. B., Martin S., Wedge D. C., Van Loo P., Ju Y. S., Smid M., Brinkman A. B., Morganella S., et al. (2016) Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47–54 10.1038/nature17676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lawrence M. S., Stojanov P., Mermel C. H., Robinson J. T., Garraway L. A., Golub T. R., Meyerson M., Gabriel S. B., Lander E. S., and Getz G. (2014) Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 10.1038/nature12912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Burns M. B., Temiz N. A., and Harris R. S. (2013) Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat. Genet. 45, 977–983 10.1038/ng.2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wijesinghe P., and Bhagwat A. S. (2012) Efficient deamination of 5-methylcytosines in DNA by human APOBEC3A, but not by AID or APOBEC3G. Nucleic Acids Res. 40, 9206–9217 10.1093/nar/gks685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wei S., Perera M. L. W., Sakhtemani R., and Bhagwat A. S. (2017) A novel class of chemicals that react with abasic sites in DNA and specifically kill B cell cancers. PLoS ONE 12, e0185010 10.1371/journal.pone.0185010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhang Z. Z., Pannunzio N. R., Hsieh C. L., Yu K., and Lieber M. R. (2015) Complexities due to single-stranded RNA during antibody detection of genomic rna:dna hybrids. BMC Res. Notes 8, 127 10.1186/s13104-015-1092-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Li H., and Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wright E. S., and Vetsigian K. H. (2016) Quality filtering of Illumina index reads mitigates sample cross-talk. BMC Genomics 17, 876 10.1186/s12864-016-3217-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. R Core Team (2013) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 78. Favorov A., Mularoni L., Cope L. M., Medvedeva Y., Mironov A. A., Makeev V. J., and Wheelan S. J. (2012) Exploring massive, genome scale datasets with the GenometriCorr package. PLoS Comput. Biol. 8, e1002529 10.1371/journal.pcbi.1002529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., Getz G., and Mesirov J. P. (2011) Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.